Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry
- PMID: 11581660
- PMCID: PMC1444949
- DOI: 10.1038/nbt1001-946
Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry
Abstract
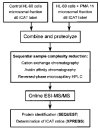
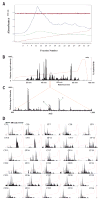
An approach to the systematic identification and quantification of the proteins contained in the microsomal fraction of cells is described. It consists of three steps: (1) preparation of microsomal fractions from cells or tissues representing different states; (2) covalent tagging of the proteins with isotope-coded affinity tag (ICAT) reagents followed by proteolysis of the combined labeled protein samples; and (3) isolation, identification, and quantification of the tagged peptides by multidimensional chromatography, automated tandem mass spectrometry, and computational analysis of the obtained data. The method was used to identify and determine the ratios of abundance of each of 491 proteins contained in the microsomal fractions of naïve and in vitro- differentiated human myeloid leukemia (HL-60) cells. The method and the new software tools to support it are well suited to the large-scale, quantitative analysis of membrane proteins and other classes of proteins that have been refractory to standard proteomics technology.
Figures


References
-
- Oseroff AR, Robbins PW, Burger MM. Cell surface membrane: biochemical aspects and biophysical probes. Annual Rev Biochem. 1973;42:647–682. - PubMed
-
- Liu E, et al. The HER2 (c-erbB-2) oncogene is frequently amplified in in situ carcinomas of the breast. Oncogene. 1992;7:1027–1032. - PubMed
-
- Shak S. Overview of the trastuzumab (Herceptin) anti-HER2 monoclonal antibody clinical program in HER2-overexpressing metastatic breast cancer. Herceptin Multinational Investigator Study Group. Semin Oncol. 1999;26:71–77. - PubMed
-
- Drews J. Research and development. Basic science and pharmaceutical innovation. Nat Biotechnol. 1999;17:406–408. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

