Antiviral actions of interferons
- PMID: 11585785
- PMCID: PMC89003
- DOI: 10.1128/CMR.14.4.778-809.2001
Antiviral actions of interferons
Abstract
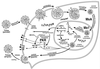
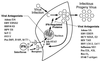
Tremendous progress has been made in understanding the molecular basis of the antiviral actions of interferons (IFNs), as well as strategies evolved by viruses to antagonize the actions of IFNs. Furthermore, advances made while elucidating the IFN system have contributed significantly to our understanding in multiple areas of virology and molecular cell biology, ranging from pathways of signal transduction to the biochemical mechanisms of transcriptional and translational control to the molecular basis of viral pathogenesis. IFNs are approved therapeutics and have moved from the basic research laboratory to the clinic. Among the IFN-induced proteins important in the antiviral actions of IFNs are the RNA-dependent protein kinase (PKR), the 2',5'-oligoadenylate synthetase (OAS) and RNase L, and the Mx protein GTPases. Double-stranded RNA plays a central role in modulating protein phosphorylation and RNA degradation catalyzed by the IFN-inducible PKR kinase and the 2'-5'-oligoadenylate-dependent RNase L, respectively, and also in RNA editing by the IFN-inducible RNA-specific adenosine deaminase (ADAR1). IFN also induces a form of inducible nitric oxide synthase (iNOS2) and the major histocompatibility complex class I and II proteins, all of which play important roles in immune response to infections. Several additional genes whose expression profiles are altered in response to IFN treatment and virus infection have been identified by microarray analyses. The availability of cDNA and genomic clones for many of the components of the IFN system, including IFN-alpha, IFN-beta, and IFN-gamma, their receptors, Jak and Stat and IRF signal transduction components, and proteins such as PKR, 2',5'-OAS, Mx, and ADAR, whose expression is regulated by IFNs, has permitted the generation of mutant proteins, cells that overexpress different forms of the proteins, and animals in which their expression has been disrupted by targeted gene disruption. The use of these IFN system reagents, both in cell culture and in whole animals, continues to provide important contributions to our understanding of the virus-host interaction and cellular antiviral response.
Figures




References
-
- Abraham N, Stojdl D F, Duncan P I, Méthot N, Ishii T, Dubé M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, Koromilas A E, Brown E G, Sonenberg N, Bell J C. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. - PubMed
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing; 1994.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

