Stress-specific activation and repression of heat shock factors 1 and 2
- PMID: 11585899
- PMCID: PMC99891
- DOI: 10.1128/MCB.21.21.7163-7171.2001
Stress-specific activation and repression of heat shock factors 1 and 2
Abstract
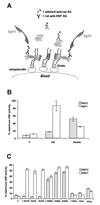
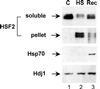
Vertebrate cells express a family of heat shock transcription factors (HSF1 to HSF4) that coordinate the inducible regulation of heat shock genes in response to diverse signals. HSF1 is potent and activated rapidly though transiently by heat shock, whereas HSF2 is a less active transcriptional regulator but can retain its DNA binding properties for extended periods. Consequently, the differential activation of HSF1 and HSF2 by various stresses may be critical for cells to survive repeated and diverse stress challenges and to provide a mechanism for more precise regulation of heat shock gene expression. Here we show, using a novel DNA binding and detection assay, that HSF1 and HSF2 are coactivated to different levels in response to a range of conditions that cause cell stress. Above a low basal activity of both HSFs, heat shock preferentially activates HSF1, whereas the amino acid analogue azetidine or the proteasome inhibitor MG132 coactivates both HSFs to different levels and hemin preferentially induces HSF2. Unexpectedly, we also found that heat shock has dramatic adverse effects on HSF2 that lead to its reversible inactivation coincident with relocalization from the nucleus. The reversible inactivation of HSF2 is specific to heat shock and does not occur with other stressors or in cells expressing high levels of heat shock proteins. These results reveal that HSF2 activity is negatively regulated by heat and suggest a role for heat shock proteins in the positive regulation of HSF2.
Figures





References
-
- Cotto J, Fox S S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110:2925–2934. - PubMed
-
- Czarnecka-Verner E, Yuan C X, Fox P C, Gurley W B. Isolation and characterization of six heat shock transcription factor cDNA clones from soybean. Plant Mol Biol. 1995;29:37–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
