Oscillating on borrowed time: diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts
- PMID: 11588167
- PMCID: PMC6763840
- DOI: 10.1523/JNEUROSCI.21-20-07937.2001
Oscillating on borrowed time: diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts
Abstract
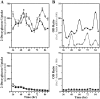
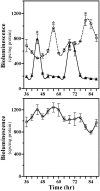
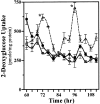
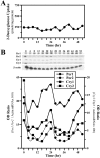
The capacity to generate circadian rhythms endogenously and to confer this rhythmicity to other cells was compared in immortalized cells derived from the suprachiasmatic nucleus (SCN) and a fibroblast line to differentiate SCN pacemaker properties from the oscillatory behavior of non-clock tissues. Only SCN2.2 cells were capable of endogenously generating circadian rhythms in 2-deoxyglucose uptake and Per gene expression. Similar to SCN function in vivo, SCN2.2 cells imposed rhythms of metabolic activity and Per gene expression on cocultured NIH/3T3 fibroblasts via a diffusible signal. The conferred rhythms in NIH/3T3 cells were phase delayed by 4-12 hr relative to SCN2.2 circadian patterns, thus resembling the phase relationship between SCN and peripheral tissue rhythms in vivo. Sustained metabolic rhythmicity in NIH/3T3 cells was dependent on continued exposure to SCN2.2-specific outputs. In response to a serum shock the NIH/3T3 fibroblasts exhibited recurrent oscillations in clock gene expression, but not in metabolic activity. These molecular rhythms in serum-shocked fibroblasts cycled in a phase relationship similar to that observed in the SCN in vivo; peak Per1 and Per2 mRNA expression preceded the rhythmic maxima in Cry1 and Cry2 mRNA levels by 4 hr. Despite these clock gene oscillations the serum-shocked NIH/3T3 cells failed to drive circadian rhythms of Per1 and Per2 expression in cocultures of untreated fibroblasts, suggesting that expression and circadian regulation of the Per and Cry genes are not sufficient to confer pacemaker function. Therefore, SCN-specific outputs are necessary to drive circadian rhythms of metabolic activity, and these output signals are not a direct product of clock gene oscillations.
Figures

References
-
- Aguilar-Roblero R, Morin LP, Moore RY. Morphological correlates of circadian rhythm restoration induced by transplantation of the suprachiasmatic nucleus in hamsters. Exp Neurol. 1994;130:250–260. - PubMed
-
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. - PubMed
-
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. - PubMed
-
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
