Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1alpha and 1gamma1
- PMID: 11588169
- PMCID: PMC6763852
- DOI: 10.1523/JNEUROSCI.21-20-07954.2001
Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1alpha and 1gamma1
Abstract
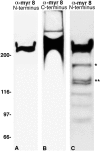
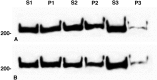
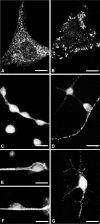
Directed neuronal, astroglial, and oligodendroglial cell migrations comprise a prominent feature of mammalian brain development. Because molecular motor proteins have been implicated in a wide spectrum of processes associated with cell motility, we initiated studies to define the pool of myosins in migrating cerebellar granule neurons and type-1 neocortical astrocytes. Our analyses identified two isoforms of a novel unconventional myosin, which we have cloned, sequenced, and designated myr 8a and 8b (eighth unconventional myosin from rat). Phylogenetic analysis indicates that myr 8 myosins comprise a new class of myosins, which we have designated class XVI. The head domain contains a large N-terminal extension composed of multiple ankyrin repeats, which are implicated in mediating an association with the protein phosphatase 1 (PP1) catalytic subunits 1alpha and 1gamma. The motor domain is followed by a single putative light-chain binding domain. The tail domain of myr 8a is comparatively short with a net positive charge, whereas the tail domain of myr 8b is extended, bears an overall neutral charge, and reveals several stretches of poly-proline residues. Neither the myr 8a nor the myr 8b sequence reveals alpha-helical coiled-coil motifs, suggesting that these myosins exist as monomers. Both immunoblot and Northern blot analyses indicate that myr 8b is the predominant isoform expressed in brain, principally at developmental time periods. The structural features and restricted expression patterns suggest that members of this novel class of unconventional myosins comprise a mechanism to target selectively the protein phosphatase 1 catalytic subunits 1alpha and/or 1gamma in developing brain.
Figures









Similar articles
-
Myr 7 is a novel myosin IX-RhoGAP expressed in rat brain.J Cell Sci. 1998 Dec 18;111 ( Pt 24):3597-608. doi: 10.1242/jcs.111.24.3597. J Cell Sci. 1998. PMID: 9819351
-
Rat myr 4 defines a novel subclass of myosin I: identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity.J Cell Biol. 1994 Jul;126(2):375-89. doi: 10.1083/jcb.126.2.375. J Cell Biol. 1994. PMID: 8034741 Free PMC article.
-
A novel mammalian myosin I from rat with an SH3 domain localizes to Con A-inducible, F-actin-rich structures at cell-cell contacts.J Cell Biol. 1995 May;129(3):819-30. doi: 10.1083/jcb.129.3.819. J Cell Biol. 1995. PMID: 7730414 Free PMC article.
-
Myosin phosphatase: subunits and interactions.Acta Physiol Scand. 1998 Dec;164(4):483-93. doi: 10.1046/j.1365-201X.1998.00447.x. Acta Physiol Scand. 1998. PMID: 9887971 Review.
-
Tails of unconventional myosins.Cell Mol Life Sci. 1999 Oct 15;56(3-4):243-57. doi: 10.1007/s000180050426. Cell Mol Life Sci. 1999. PMID: 11212352 Free PMC article. Review.
Cited by
-
NELF is a nuclear protein involved in hypothalamic GnRH neuronal migration.Mol Cell Endocrinol. 2010 May 5;319(1-2):47-55. doi: 10.1016/j.mce.2009.11.016. Epub 2009 Dec 16. Mol Cell Endocrinol. 2010. PMID: 20025934 Free PMC article.
-
Signal transduction therapeutics: relevance for Alzheimer's disease.J Mol Neurosci. 2004;23(1-2):123-42. doi: 10.1385/JMN:23:1-2:123. J Mol Neurosci. 2004. PMID: 15126698 Review.
-
The C-terminal tail extension of myosin 16 acts as a molten globule, including intrinsically disordered regions, and interacts with the N-terminal ankyrin.J Biol Chem. 2021 Jul;297(1):100716. doi: 10.1016/j.jbc.2021.100716. Epub 2021 Apr 28. J Biol Chem. 2021. PMID: 33930467 Free PMC article.
-
NYAP: a phosphoprotein family that links PI3K to WAVE1 signalling in neurons.EMBO J. 2011 Sep 23;30(23):4739-54. doi: 10.1038/emboj.2011.348. EMBO J. 2011. PMID: 21946561 Free PMC article.
-
Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals.Transl Psychiatry. 2017 May 9;7(5):e1119. doi: 10.1038/tp.2017.93. Transl Psychiatry. 2017. PMID: 28485726 Free PMC article.
References
-
- Adams RJ, Pollard TD. Binding of myosin I to membrane lipids. Nature. 1989;340:565–568. - PubMed
-
- Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure and function. CRC; Boca Raton, FL: 1997.
-
- Andersen SSL, Bi GQ. Axon formation: a molecular model for generation of neuronal polarity. BioEssays. 2000;22:172–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
