Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence
- PMID: 11591641
- PMCID: PMC311145
- DOI: 10.1101/gr.190201
Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence
Abstract
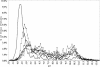
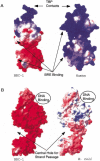
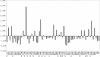
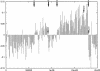
The genome of the halophilic archaeon Halobacterium sp. NRC-1 and predicted proteome have been analyzed by computational methods and reveal characteristics relevant to life in an extreme environment distinguished by hypersalinity and high solar radiation: (1) The proteome is highly acidic, with a median pI of 4.9 and mostly lacking basic proteins. This characteristic correlates with high surface negative charge, determined through homology modeling, as the major adaptive mechanism of halophilic proteins to function in nearly saturating salinity. (2) Codon usage displays the expected GC bias in the wobble position and is consistent with a highly acidic proteome. (3) Distinct genomic domains of NRC-1 with bacterial character are apparent by whole proteome BLAST analysis, including two gene clusters coding for a bacterial-type aerobic respiratory chain. This result indicates that the capacity of halophiles for aerobic respiration may have been acquired through lateral gene transfer. (4) Two regions of the large chromosome were found with relatively lower GC composition and overrepresentation of IS elements, similar to the minichromosomes. These IS-element-rich regions of the genome may serve to exchange DNA between the three replicons and promote genome evolution. (5) GC-skew analysis showed evidence for the existence of two replication origins in the large chromosome. This finding and the occurrence of multiple chromosomes indicate a dynamic genome organization with eukaryotic character.
Figures



Similar articles
-
Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1.BMC Genet. 2007 Jun 8;8:31. doi: 10.1186/1471-2156-8-31. BMC Genet. 2007. PMID: 17559652 Free PMC article.
-
Low-pass sequencing for microbial comparative genomics.BMC Genomics. 2004 Jan 12;5(1):3. doi: 10.1186/1471-2164-5-3. BMC Genomics. 2004. PMID: 14718067 Free PMC article.
-
Genome sequence of Halobacterium species NRC-1.Proc Natl Acad Sci U S A. 2000 Oct 24;97(22):12176-81. doi: 10.1073/pnas.190337797. Proc Natl Acad Sci U S A. 2000. PMID: 11016950 Free PMC article.
-
Bacterial genomic G+C composition-eliciting environmental adaptation.Genomics. 2010 Jan;95(1):7-15. doi: 10.1016/j.ygeno.2009.09.002. Epub 2009 Sep 9. Genomics. 2010. PMID: 19747541 Review.
-
Halophiles and their enzymes: negativity put to good use.Curr Opin Microbiol. 2015 Jun;25:120-6. doi: 10.1016/j.mib.2015.05.009. Epub 2015 Jun 9. Curr Opin Microbiol. 2015. PMID: 26066288 Free PMC article. Review.
Cited by
-
The core and unique proteins of haloarchaea.BMC Genomics. 2012 Jan 24;13:39. doi: 10.1186/1471-2164-13-39. BMC Genomics. 2012. PMID: 22272718 Free PMC article.
-
The origin and evolution of Archaea: a state of the art.Philos Trans R Soc Lond B Biol Sci. 2006 Jun 29;361(1470):1007-22. doi: 10.1098/rstb.2006.1841. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 16754611 Free PMC article. Review.
-
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.BMC Genomics. 2007 Mar 29;8:86. doi: 10.1186/1471-2164-8-86. BMC Genomics. 2007. PMID: 17394648 Free PMC article.
-
Molecular Adaptations of Bacterial Mercuric Reductase to the Hypersaline Kebrit Deep in the Red Sea.Appl Environ Microbiol. 2019 Feb 6;85(4):e01431-18. doi: 10.1128/AEM.01431-18. Print 2019 Feb 15. Appl Environ Microbiol. 2019. PMID: 30504211 Free PMC article.
-
Double mutations far from the active site affect cold activity in an Antarctic halophilic β-galactosidase.Protein Sci. 2022 Mar;31(3):677-687. doi: 10.1002/pro.4264. Epub 2022 Jan 5. Protein Sci. 2022. PMID: 34939242 Free PMC article.
References
-
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol. 2000;36:1184–1185. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
-
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. ; erratum 380: 179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
