A comparative molecular analysis of developing mouse forelimbs and hindlimbs using serial analysis of gene expression (SAGE)
- PMID: 11591645
- PMCID: PMC311149
- DOI: 10.1101/gr.192601
A comparative molecular analysis of developing mouse forelimbs and hindlimbs using serial analysis of gene expression (SAGE)
Abstract
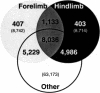
The analysis of differentially expressed genes is a powerful approach to elucidate the genetic mechanisms underlying the morphological and evolutionary diversity among serially homologous structures, both within the same organism (e.g., hand vs. foot) and between different species (e.g., hand vs. wing). In the developing embryo, limb-specific expression of Pitx1, Tbx4, and Tbx5 regulates the determination of limb identity. However, numerous lines of evidence, including the fact that these three genes encode transcription factors, indicate that additional genes are involved in the Pitx1-Tbx hierarchy. To examine the molecular distinctions coded for by these factors, and to identify novel genes involved in the determination of limb identity, we have used Serial Analysis of Gene Expression (SAGE) to generate comprehensive gene expression profiles from intact, developing mouse forelimbs and hindlimbs. To minimize the extraction of erroneous SAGE tags from low-quality sequence data, we used a new algorithm to extract tags from -analyzed sequence data and obtained 68,406 and 68,450 SAGE tags from forelimb and hindlimb SAGE libraries, respectively. We also developed an improved method for determining the identity of SAGE tags that increases the specificity of and provides additional information about the confidence of the tag-UniGene cluster match. The most differentially expressed gene between our SAGE libraries was Pitx1. The differential expression of Tbx4, Tbx5, and several limb-specific Hox genes was also detected; however, their abundances in the SAGE libraries were low. Because numerous other tags were differentially expressed at this low level, we performed a 'virtual' subtraction with 362,344 tags from six additional nonlimb SAGE libraries to further refine this set of candidate genes. This subtraction reduced the number of candidate genes by 74%, yet preserved the previously identified regulators of limb identity. This study presents the gene expression complexity of the developing limb and identifies candidate genes involved in the regulation of limb identity. We propose that our computational tools and the overall strategy used here are broadly applicable to other SAGE-based studies in a variety of organisms. [SAGE data are all available at GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSM55 and GSM56, which correspond to the forelimb and hindlimb raw SAGE data.]
Figures





Similar articles
-
A comparative analysis of the information content in long and short SAGE libraries.BMC Bioinformatics. 2006 Nov 16;7:504. doi: 10.1186/1471-2105-7-504. BMC Bioinformatics. 2006. PMID: 17109755 Free PMC article.
-
How the embryo makes a limb: determination, polarity and identity.J Anat. 2015 Oct;227(4):418-30. doi: 10.1111/joa.12361. Epub 2015 Aug 7. J Anat. 2015. PMID: 26249743 Free PMC article. Review.
-
Differentially expressed genes in pancreatic ductal adenocarcinomas identified through serial analysis of gene expression.Cancer Biol Ther. 2004 Dec;3(12):1254-61. doi: 10.4161/cbt.3.12.1238. Epub 2004 Dec 14. Cancer Biol Ther. 2004. PMID: 15477757
-
SAGE profiling of the forelimb and hindlimb.Genome Biol. 2002;3(3):REVIEWS1007. doi: 10.1186/gb-2002-3-3-reviews1007. Epub 2002 Feb 27. Genome Biol. 2002. PMID: 11897030 Free PMC article. Review.
-
The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity.Nature. 1999 Apr 29;398(6730):814-8. doi: 10.1038/19769. Nature. 1999. PMID: 10235264
Cited by
-
A serial analysis of gene expression profile of the Alzheimer's disease Tg2576 mouse model.Neurotox Res. 2010 May;17(4):360-79. doi: 10.1007/s12640-009-9112-3. Epub 2009 Sep 16. Neurotox Res. 2010. PMID: 19760337
-
Lineage-specific responses to reduced embryonic Pax3 expression levels.Dev Biol. 2008 Mar 15;315(2):369-82. doi: 10.1016/j.ydbio.2007.12.020. Epub 2007 Dec 27. Dev Biol. 2008. PMID: 18243171 Free PMC article.
-
Identification of novel transcripts with differential dorso-ventral expression in Xenopus gastrula using serial analysis of gene expression.Genome Biol. 2009 Feb 11;10(2):R15. doi: 10.1186/gb-2009-10-2-r15. Genome Biol. 2009. PMID: 19210784 Free PMC article.
-
Clustering of molecular alterations in gastroesophageal carcinomas.Neoplasia. 2004 Mar-Apr;6(2):143-9. doi: 10.1593/neo.03385. Neoplasia. 2004. PMID: 15140403 Free PMC article.
-
Anatomical Network Comparison of Human Upper and Lower, Newborn and Adult, and Normal and Abnormal Limbs, with Notes on Development, Pathology and Limb Serial Homology vs. Homoplasy.PLoS One. 2015 Oct 9;10(10):e0140030. doi: 10.1371/journal.pone.0140030. eCollection 2015. PLoS One. 2015. PMID: 26452269 Free PMC article.
References
-
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. - PubMed
-
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. - PubMed
-
- Capdevila J, Izpisúa-Belmonte JC. Perspectives on the evolutionary origin of tetrapod limbs. J Exp Zool. 2000;288:287–303. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
