Identification of the partitioning site within the repABC-type replicon of the composite Paracoccus versutus plasmid pTAV1
- PMID: 11591666
- PMCID: PMC100104
- DOI: 10.1128/JB.183.21.6234-6243.2001
Identification of the partitioning site within the repABC-type replicon of the composite Paracoccus versutus plasmid pTAV1
Abstract
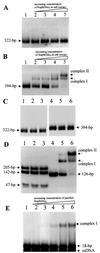
The replicator region of composite plasmid pTAV1 of Paracoccus versutus (included in mini-replicon pTAV320) belongs to the family of repABC replicons commonly found in plasmids harbored by Agrobacterium and Rhizobium spp. The repABC replicons encode three genes clustered in an operon, which are involved in partitioning (repA and repB) and replication (repC). In order to localize the partitioning site of pTAV320, the two identified incompatibility determinants of this mini-replicon (inc1, located in the intergenic sequence between repB and repC; and inc2, situated downstream of the repC gene) were PCR amplified and used together with purified RepB fusion protein (homologous to the type B partitioning proteins binding to the partitioning sites) in an electrophoretic mobility shift assay. The protein bound only inc2, forming two complexes in a protein concentration-dependent manner. The inc2 region contains two long (14-bp) repeated sequences (R1 and R2). Disruption of these sequences completely eliminates RepB binding ability. R1 and R2 have sequence similarities with analogous repeats of another repABC replicon of plasmid pPAN1 of Paracoccus pantotrophus DSM 82.5 and with centromeric sequences of the Bacillus subtilis chromosome. Excess RepB protein resulted in destabilization of the inc2-containing plasmid in Escherichia coli. On the other hand, the inc2 region could stabilize another unstable replicon in P. versutus when RepA and RepB were delivered in trans, proving that this region has centromere-like activity. Thus, it was demonstrated that repA, repB, and inc2 constitute a functional system for active partitioning of pTAV320.
Figures





References
-
- Austin S, Abeles A L. Partitioning of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. - PubMed
-
- Austin S, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. - PubMed
-
- Bartosik D, Baj J, Wlodarczyk M. Construction and preliminary characterization of mini-derivatives of large (107-kb) cryptic plasmid of the sulphur bacterium Thiobacillus versutus. FEMS Microbiol Lett. 1995;129:169–174.
-
- Bartosik D, Bialkowska A, Baj J, Wlodarczyk M. Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol Pol. 1997;46:379–383. - PubMed
-
- Bartosik D, Baj J, Wlodarczyk M. Molecular and functional analysis of pTAV320-repABC type replicon of a composite pTAV1 plasmid of Paracoccus versutus. Microbiology. 1998;144:3149–3157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

