Regulation of rRNA transcription correlates with nucleoside triphosphate sensing
- PMID: 11591676
- PMCID: PMC100125
- DOI: 10.1128/JB.183.21.6315-6323.2001
Regulation of rRNA transcription correlates with nucleoside triphosphate sensing
Abstract
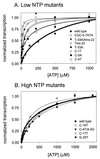
We have previously shown that the activity of the Escherichia coli rRNA promoter rrnB P1 in vitro depends on the concentration of the initiating nucleotide, ATP, and can respond to changes in ATP pools in vivo. We have proposed that this nucleoside triphosphate (NTP) sensing might contribute to regulation of rRNA transcription. To test this model, we have measured the ATP requirements for transcription from 11 different rrnB P1 core promoter mutants in vitro and compared them with the regulatory responses of the same promoters in vivo. The seven rrnB P1 variants that required much lower ATP concentrations than the wild-type promoter for efficient transcription in vitro were defective for response to growth rate changes in vivo (growth rate-dependent regulation). In contrast, the four variants requiring high ATP concentrations in vitro (like the wild-type promoter) were regulated with the growth rate in vivo. We also observed a correlation between NTP sensing in vitro and the response of the promoters in vivo to deletion of the fis gene (an example of homeostatic control), although this relationship was not as tight as for growth rate-dependent regulation. We conclude that the kinetic features responsible for the high ATP concentration dependence of the rrnB P1 promoter in vitro are responsible, at least in part, for the promoter's regulation in vivo, consistent with the model in which rrnB P1 promoter activity can be regulated by changes in NTP pools in vivo (or by hypothetical factors that work at the same kinetic steps that make the promoter sensitive to NTPs).
Figures



References
-
- Afflerbach H, Schroder O, Wagner R. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 1998;28:641–653. - PubMed
-
- Barker M M, Gaal T, Gourse R L. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. - PubMed
-
- Barker M M, Gaal T, Josaitis C A, Gourse R L. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

