Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA
- PMID: 11593008
- PMCID: PMC59823
- DOI: 10.1073/pnas.211266898
Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA
Abstract
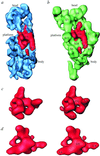
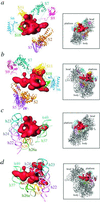
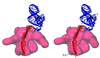
S1 is the largest ribosomal protein, present in the small subunit of the bacterial ribosome. It has a pivotal role in stabilizing the mRNA on the ribosome. Thus far, S1 has eluded structural determination. We have identified the S1 protein mass in the cryo-electron microscopic map of the Escherichia coli ribosome by comparing the map with a recent x-ray crystallographic structure of the 30S subunit, which lacks S1. According to our finding, S1 is located at the junction of head, platform, and main body of the 30S subunit, thus explaining all existing biochemical and crosslinking data. Protein S1 as identified in our map has a complex, elongated shape with two holes in its central portion. The N-terminal domain, forming one of the extensions, penetrates into the head of the 30S subunit. Evidence for direct interaction of S1 with 11 nucleotides of the mRNA, immediately upstream of the Shine-Dalgarno sequence, explains the protein's role in the recognition of the 5' region of mRNA.
Figures

Similar articles
-
All-atom homology model of the Escherichia coli 30S ribosomal subunit.Nat Struct Biol. 2002 Oct;9(10):750-5. doi: 10.1038/nsb841. Nat Struct Biol. 2002. PMID: 12244297
-
Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly.Mol Cell. 2005 Apr 29;18(3):319-29. doi: 10.1016/j.molcel.2005.03.028. Mol Cell. 2005. PMID: 15866174
-
How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.Science. 2012 May 18;336(6083):915-8. doi: 10.1126/science.1218538. Science. 2012. PMID: 22605777 Free PMC article.
-
The ribosome through the looking glass.Angew Chem Int Ed Engl. 2003 Aug 4;42(30):3464-86. doi: 10.1002/anie.200200544. Angew Chem Int Ed Engl. 2003. PMID: 12900959 Review.
-
How does the mRNA pass through the ribosome?Biochimie. 1991 Jul-Aug;73(7-8):937-45. doi: 10.1016/0300-9084(91)90135-n. Biochimie. 1991. PMID: 1742365 Review.
Cited by
-
Identification of an AU-rich translational enhancer within the Escherichia coli fepB leader RNA.J Bacteriol. 2007 Jun;189(11):4028-37. doi: 10.1128/JB.01924-06. Epub 2007 Mar 30. J Bacteriol. 2007. PMID: 17400738 Free PMC article.
-
Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM.Methods. 2018 Mar 15;137:55-66. doi: 10.1016/j.ymeth.2017.12.004. Epub 2017 Dec 14. Methods. 2018. PMID: 29247757 Free PMC article.
-
Automated design of synthetic ribosome binding sites to control protein expression.Nat Biotechnol. 2009 Oct;27(10):946-50. doi: 10.1038/nbt.1568. Epub 2009 Oct 4. Nat Biotechnol. 2009. PMID: 19801975 Free PMC article.
-
Crosslinking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh.RNA. 2002 May;8(5):612-25. doi: 10.1017/s1355838202020095. RNA. 2002. PMID: 12022228 Free PMC article.
-
Global analysis of protein synthesis in Flavobacterium johnsoniae reveals the use of Kozak-like sequences in diverse bacteria.Nucleic Acids Res. 2019 Nov 18;47(20):10477-10488. doi: 10.1093/nar/gkz855. Nucleic Acids Res. 2019. PMID: 31602466 Free PMC article.
References
-
- Wittmann H G. In: Ribosomes. Namura M, Tissieres A, Lengyel P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. pp. 93–114.
-
- Subramanian A R. Trends Biochem Sci. 1984;9:491–494.
-
- Tzareva N V, Makhno V I, Boni I V. FEBS Lett. 1994;337:189–194. - PubMed
-
- Potapov A P, Subramanian A R. Biochem Int. 1992;27:745–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

