Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis
- PMID: 11593010
- PMCID: PMC59757
- DOI: 10.1073/pnas.211229198
Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis
Abstract
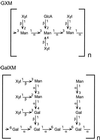
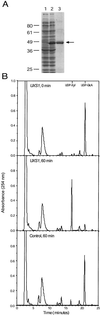
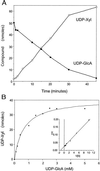
UDP-xylose is a sugar donor required for the synthesis of diverse and important glycan structures in animals, plants, fungi, and bacteria. Xylose-containing glycans are particularly abundant in plants and in the polysaccharide capsule that is the major virulence factor of the pathogenic fungus Cryptococcus neoformans. Biosynthesis of UDP-xylose is mediated by UDP-glucuronic acid decarboxylase, which converts UDP-glucuronic acid to UDP-xylose. Although this enzymatic activity was described over 40 years ago it has never been fully purified, and the gene encoding it has not been identified. We used homology to a bacterial gene, hypothesized to encode a related function, to identify a cryptococcal sequence as putatively encoding a UDP-glucuronic acid decarboxylase. A soluble 47-kDa protein derived from bacteria expressing the C. neoformans gene catalyzed conversion of UDP-glucuronic acid to UDP-xylose, as confirmed by NMR analysis. NADH, UDP, and UDP-xylose inhibit the activity. Close homologs of the cryptococcal gene, which we termed UXS1, appear in genome sequence data from organisms ranging from bacteria to humans.
Figures
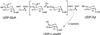


References
-
- Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, DC: Am. Soc. Microbiol.; 1998.
-
- Rodrigues M L, Alviano C S, Travassos L R. Microbes Infection. 1999;1:293–301. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

