The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome
- PMID: 11593022
- PMCID: PMC59775
- DOI: 10.1073/pnas.201182798
The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome
Erratum in
- Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):414. Melo, M [corrected to Melo, MB]
Abstract
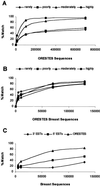
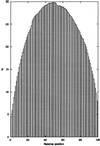
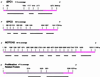
Open reading frame expressed sequences tags (ORESTES) differ from conventional ESTs by providing sequence data from the central protein coding portion of transcripts. We generated a total of 696,745 ORESTES sequences from 24 human tissues and used a subset of the data that correspond to a set of 15,095 full-length mRNAs as a means of assessing the efficiency of the strategy and its potential contribution to the definition of the human transcriptome. We estimate that ORESTES sampled over 80% of all highly and moderately expressed, and between 40% and 50% of rarely expressed, human genes. In our most thoroughly sequenced tissue, the breast, the 130,000 ORESTES generated are derived from transcripts from an estimated 70% of all genes expressed in that tissue, with an equally efficient representation of both highly and poorly expressed genes. In this respect, we find that the capacity of the ORESTES strategy both for gene discovery and shotgun transcript sequence generation significantly exceeds that of conventional ESTs. The distribution of ORESTES is such that many human transcripts are now represented by a scaffold of partial sequences distributed along the length of each gene product. The experimental joining of the scaffold components, by reverse transcription-PCR, represents a direct route to transcript finishing that may represent a useful alternative to full-length cDNA cloning.
Figures

Comment in
-
Navigating the human transcriptome.Proc Natl Acad Sci U S A. 2001 Oct 9;98(21):11837-8. doi: 10.1073/pnas.221463598. Proc Natl Acad Sci U S A. 2001. PMID: 11592992 Free PMC article. Review. No abstract available.
References
-
- Dunham I, Shimizu N, Roe B A, Chissoe S, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, Smink L J, et al. Nature (London) 1999;402:489–495. - PubMed
-
- Claverie J M. Hum Mol Genet. 1997;6:1735–1744. - PubMed
-
- Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. - PubMed
-
- Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

