Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster
- PMID: 11593024
- PMCID: PMC59777
- DOI: 10.1073/pnas.201413098
Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster
Abstract
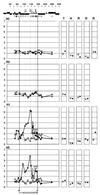
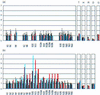
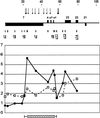
We have analyzed the pattern of core histone acetylation across 250 kb of the telomeric region of the short arm of human chromosome 16. This gene-dense region, which includes the alpha-globin genes and their regulatory elements embedded within widely expressed genes, shows marked differences in histone acetylation between erythroid and non-erythroid cells. In non-erythroid cells, there was a uniform 2- to 3-fold enrichment of acetylated histones, compared with heterochromatin, across the entire region. In erythroid cells, an approximately 100-kb segment of chromatin encompassing the alpha genes and their remote major regulatory element was highly enriched in histone H4 acetylated at Lys-5. Other lysines in the N-terminal tail of histone H4 showed intermediate and variable levels of enrichment. Similar broad segments of erythroid-specific histone acetylation were found in the corresponding syntenic regions containing the mouse and chicken alpha-globin gene clusters. The borders of these regions of acetylation are located in similar positions in all three species, and a sharply defined 3' boundary coincides with the previously identified breakpoint in conserved synteny between these species. We have therefore demonstrated that an erythroid-specific domain of acetylation has been conserved across several species, encompassing not only the alpha-globin genes but also a neighboring widely expressed gene. These results contrast with those at other clusters and demonstrate that not all genes are organized into discrete regulatory domains.
Figures


References
-
- Wolffe A, editor. Chromatin: Structure and Function. London: Academic; 1995.
-
- Bell A C, Felsenfeld G. Curr Opin Genet Dev. 1999;9:191–198. - PubMed
-
- Bell A C, West A G, Felsenfeld G. Science. 2001;291:447–450. - PubMed
-
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett N A, King A, Higgs D R. Nat Genet. 1997;15:252–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

