Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching
- PMID: 11593039
- PMCID: PMC59793
- DOI: 10.1073/pnas.221289898
Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching
Abstract
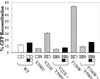
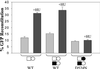
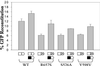
We recently proposed a dynamic copy-choice model for retroviral recombination in which a steady state between the rates of polymerization and RNA degradation determines the frequency of reverse transcriptase (RT) template switching. The relative contributions of polymerase-dependent and polymerase-independent RNase H activities during reverse transcription and template switching in vivo have not been determined. We developed an in vivo trans-complementation assay in which direct repeat deletion through template switching reconstitutes a functional green fluorescent protein gene in a retroviral vector. Complementation in trans between murine leukemia virus Gag-Pol proteins lacking polymerase and RNase H activities restored viral replication. Because only polymerase-independent RNase H activity is present in this cell line, the relative roles of polymerase-dependent and -independent RNase H activities in template switching could be determined. We also analyzed double mutants possessing polymerase and RNase H mutations that increased and decreased template switching, respectively. The double mutants exhibited low template switching frequency, indicating that the RNase H mutations were dominant. Trans-complementation of the double mutants with polymerase-independent RNase H did not restore the high template switching frequency, indicating that polymerase-dependent RNase H activity was essential for the increased frequency of template switching. Additionally, trans-complementation of RNase H mutants in the presence and absence of hydroxyurea, which slows the rate of reverse transcription, showed that hydroxyurea increased template switching only when polymerase-dependent RNase H activity was present. This is, to our knowledge, the first demonstration of polymerase-dependent RNase H activity in vivo. These results provide strong evidence for a dynamic association between the rates of DNA polymerization and polymerase-dependent RNase H activity, which determines the frequency of in vivo template switching.
Figures



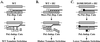
Similar articles
-
Mutations in the RNase H primer grip domain of murine leukemia virus reverse transcriptase decrease efficiency and accuracy of plus-strand DNA transfer.J Virol. 2005 Jan;79(1):419-27. doi: 10.1128/JVI.79.1.419-427.2005. J Virol. 2005. PMID: 15596835 Free PMC article.
-
Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching.J Virol. 2000 Aug;74(15):7171-8. doi: 10.1128/jvi.74.15.7171-7178.2000. J Virol. 2000. PMID: 10888659 Free PMC article.
-
Y586F mutation in murine leukemia virus reverse transcriptase decreases fidelity of DNA synthesis in regions associated with adenine-thymine tracts.Proc Natl Acad Sci U S A. 2002 Jul 23;99(15):10090-5. doi: 10.1073/pnas.152186199. Epub 2002 Jul 15. Proc Natl Acad Sci U S A. 2002. PMID: 12119402 Free PMC article.
-
The involvement of HIV-1 RNAse H in resistance to nucleoside analogues.J Antimicrob Chemother. 2008 May;61(5):973-5. doi: 10.1093/jac/dkn060. Epub 2008 Mar 5. J Antimicrob Chemother. 2008. PMID: 18325896 Review.
-
A polymerase-site-jumping model for strand transfer during DNA synthesis by reverse transcriptase.Virus Res. 2009 Sep;144(1-2):65-73. doi: 10.1016/j.virusres.2009.03.022. Epub 2009 Apr 10. Virus Res. 2009. PMID: 19427048 Review.
Cited by
-
Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor.J Virol. 2005 Apr;79(7):4140-9. doi: 10.1128/JVI.79.7.4140-4149.2005. J Virol. 2005. PMID: 15767415 Free PMC article.
-
Pausing during reverse transcription increases the rate of retroviral recombination.J Virol. 2006 Mar;80(5):2483-94. doi: 10.1128/JVI.80.5.2483-2494.2006. J Virol. 2006. PMID: 16474155 Free PMC article.
-
A Conserved Stem-Loop Structure within ORF5 Is a Frequent Recombination Hotspot for Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) with a Particular Modified Live Virus (MLV) Strain.Viruses. 2023 Jan 16;15(1):258. doi: 10.3390/v15010258. Viruses. 2023. PMID: 36680298 Free PMC article.
-
HIV-1 Ribonuclease H: Structure, Catalytic Mechanism and Inhibitors.Viruses. 2010 Apr;2(4):900-926. doi: 10.3390/v2040900. Epub 2010 Mar 30. Viruses. 2010. PMID: 21994660 Free PMC article.
-
Dynamics of HIV-1 recombination in its natural target cells.Proc Natl Acad Sci U S A. 2004 Mar 23;101(12):4204-9. doi: 10.1073/pnas.0306764101. Epub 2004 Mar 9. Proc Natl Acad Sci U S A. 2004. PMID: 15010526 Free PMC article.
References
-
- Schinazi R F, Larder B A, Mellors J W. Int Antiviral News. 2000;8:65–91.
-
- McKeating J A, Gow J, Goudsmit J, Pearl L H, Mulder C, Weiss R A. AIDS. 1989;3:777–784. - PubMed
-
- Phillips R E, Rowland-Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R, Rizza C R, et al. Nature (London) 1991;354:453–459. - PubMed
-
- Reitz M S, Wilson C, Naugle C, Gallo R C, Robert-Guroff M. Cell. 1988;54:57–63. - PubMed
-
- Pathak V K, Hu W S. Semin Virol. 1997;8:141–150.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

