Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression
- PMID: 11595803
- PMCID: PMC139160
- DOI: 10.1105/tpc.010240
Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression
Abstract
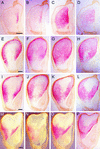
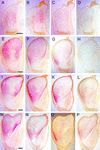
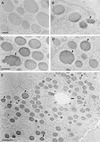
We analyzed cDNA libraries from developing endosperm of the B73 maize inbred line to evaluate the expression of storage protein genes. This study showed that zeins are by far the most highly expressed genes in the endosperm, but we found an inverse relationship between the number of zein genes and the relative amount of specific mRNAs. Although alpha-zeins are encoded by large multigene families, only a few of these genes are transcribed at high or detectable levels. In contrast, relatively small gene families encode the gamma- and delta-zeins, and members of these gene families, especially the gamma-zeins, are highly expressed. Knowledge of expressed storage protein genes allowed the development of DNA and antibody probes that distinguish between closely related gene family members. Using in situ hybridization, we found differences in the temporal and spatial expression of the alpha-, gamma-, and delta-zein gene families, which provides evidence that gamma-zeins are synthesized throughout the endosperm before alpha- and delta-zeins. This observation is consistent with earlier studies that suggested that gamma-zeins play an important role in prolamin protein body assembly. Analysis of endosperm cDNAs also revealed several previously unidentified proteins, including a 50-kD gamma-zein, an 18-kD alpha-globulin, and a legumin-related protein. Immunolocalization of the 50-kD gamma-zein showed this protein to be located at the surface of prolamin-containing protein bodies, similar to other gamma-zeins. The 18-kD alpha-globulin, however, is deposited in novel, vacuole-like organelles that were not described previously in maize endosperm.
Figures






Similar articles
-
Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm.Plant Cell. 2002 Mar;14(3):655-72. doi: 10.1105/tpc.010431. Plant Cell. 2002. PMID: 11910012 Free PMC article.
-
Allelic variation and differential expression of methionine-rich delta-zeins in maize inbred lines B73 and W23a1.Planta. 2003 May;217(1):66-74. doi: 10.1007/s00425-002-0971-6. Epub 2003 Feb 20. Planta. 2003. PMID: 12721850
-
Maize opaque10 Encodes a Cereal-Specific Protein That Is Essential for the Proper Distribution of Zeins in Endosperm Protein Bodies.PLoS Genet. 2016 Aug 19;12(8):e1006270. doi: 10.1371/journal.pgen.1006270. eCollection 2016 Aug. PLoS Genet. 2016. Retraction in: PLoS Genet. 2022 Nov 16;18(11):e1010501. doi: 10.1371/journal.pgen.1010501. PMID: 27541862 Free PMC article. Retracted.
-
The regulation of zein biosynthesis in maize endosperm.Theor Appl Genet. 2020 May;133(5):1443-1453. doi: 10.1007/s00122-019-03520-z. Epub 2020 Jan 2. Theor Appl Genet. 2020. PMID: 31897513 Review.
-
Structural elements regulating zein gene expression.Bioessays. 1989 Apr;10(4):108-13. doi: 10.1002/bies.950100404. Bioessays. 1989. PMID: 2658986 Review.
Cited by
-
Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm.Proc Natl Acad Sci U S A. 2013 May 7;110(19):E1827-36. doi: 10.1073/pnas.1304903110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610440 Free PMC article.
-
Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm.Plant Physiol. 2013 Jul;162(3):1359-69. doi: 10.1104/pp.113.218941. Epub 2013 May 15. Plant Physiol. 2013. PMID: 23677936 Free PMC article.
-
Allelic variation of the β-, γ- and δ-kafirin genes in diverse Sorghum genotypes.Theor Appl Genet. 2010 Nov;121(7):1227-37. doi: 10.1007/s00122-010-1383-9. Epub 2010 Jun 19. Theor Appl Genet. 2010. PMID: 20563549
-
Maize opaque endosperm mutations create extensive changes in patterns of gene expression.Plant Cell. 2002 Oct;14(10):2591-612. doi: 10.1105/tpc.003905. Plant Cell. 2002. PMID: 12368507 Free PMC article.
-
Duplication-dependent CG suppression of the seed storage protein genes of maize.Genetics. 2003 Oct;165(2):835-48. doi: 10.1093/genetics/165.2.835. Genetics. 2003. PMID: 14573492 Free PMC article.
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 21, 5403–5410. - PubMed
-
- Benner, M., Phillips, R.L., Kirihara, J.A., and Messing, J. (1989). Genetic analysis of methionine-rich storage protein accumulation in maize. Theor. Appl. Genet. 78, 761–767. - PubMed
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. - PubMed
-
- Burr, B., and Burr, F.A. (1991). Recombinant inbreds for molecular mapping in maize: Theoretical and practical considerations. Trends Genet. 7, 55–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

