The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria
- PMID: 11597336
- PMCID: PMC57799
- DOI: 10.1186/gb-2001-2-10-research0044
The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria
Abstract
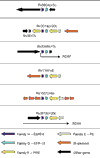
Background: The genome of Mycobacterium tuberculosis H37Rv has five copies of a cluster of genes known as the ESAT-6 loci. These clusters contain members of the CFP-10 (lhp) and ESAT-6 (esat-6) gene families (encoding secreted T-cell antigens that lack detectable secretion signals) as well as genes encoding secreted, cell-wall-associated subtilisin-like serine proteases, putative ABC transporters, ATP-binding proteins and other membrane-associated proteins. These membrane-associated and energy-providing proteins may function to secrete members of the ESAT-6 and CFP-10 protein families, and the proteases may be involved in processing the secreted peptide.
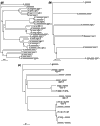
Results: Finished and unfinished genome sequencing data of 98 publicly available microbial genomes has been analyzed for the presence of orthologs of the ESAT-6 loci. The multiple duplicates of the ESAT-6 gene cluster found in the genome of M. tuberculosis H37Rv are also conserved in the genomes of other mycobacteria, for example M. tuberculosis CDC1551, M. tuberculosis 210, M. bovis, M. leprae, M. avium, and the avirulent strain M. smegmatis. Phylogenetic analyses of the resulting sequences have established the duplication order of the gene clusters and demonstrated that the gene cluster known as region 4 (Rv3444c-3450c) is ancestral. Region 4 is also the only region for which an ortholog could be found in the genomes of Corynebacterium diphtheriae and Streptomyces coelicolor.
Conclusions: Comparative genomic analysis revealed that the presence of the ESAT-6 gene cluster is a feature of some high-G+C Gram-positive bacteria. Multiple duplications of this cluster have occurred and are maintained only within the genomes of members of the genus Mycobacterium.
Figures





References
-
- Philipp WJ, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole ST. Physical mapping of Mycobacterium bovis BCG pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 1996;142:3135–3145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

