Having a BLAST with bioinformatics (and avoiding BLASTphemy)
- PMID: 11597340
- PMCID: PMC138974
- DOI: 10.1186/gb-2001-2-10-reviews2002
Having a BLAST with bioinformatics (and avoiding BLASTphemy)
Abstract
Searching for similarities between biological sequences is the principal means by which bioinformatics contributes to our understanding of biology. Of the various informatics tools developed to accomplish this task, the most widely used is BLAST, the basic local alignment search tool. This article discusses the principles, workings, applications and potential pitfalls of BLAST, focusing on the implementation developed at the National Center for Biotechnology Information.
Figures

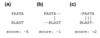

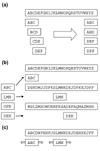


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- NCBI BLASt http://www.ncbi.nlm.nih.gov/BLAST/
-
- WU-BLAST http://blast.wustl.edu/
-
- Baxevanis AD, Ouellette BFF, (eds) Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins John Wiley; 1998.
-
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids Cambridge: Cambridge University Press; 1998.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

