Exploration of the pore structure of a peptide-gated Na+ channel
- PMID: 11598003
- PMCID: PMC125683
- DOI: 10.1093/emboj/20.20.5595
Exploration of the pore structure of a peptide-gated Na+ channel
Abstract
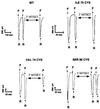
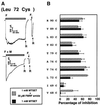
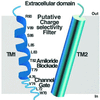
The FMRF-amide-activated sodium channel (FaNaC), a member of the ENaC/Degenerin family, is a homotetramer, each subunit containing two transmembrane segments. We changed independently every residue of the first transmembrane segment (TM1) into a cysteine and tested each position's accessibility to the cysteine covalent reagents MTSET and MTSES. Eleven mutants were accessible to the cationic MTSET, showing that TM1 faces the ion translocation pathway. This was confirmed by the accessibility of cysteines present in the acid-sensing ion channels and other mutations introduced in FaNaC TM1. Modification of accessibilities for positions 69, 71 and 72 in the open state shows that the gating mechanism consists of the opening of a constriction close to the intracellular side. The anionic MTSES did not penetrate into the channel, indicating the presence of a charge selectivity filter in the outer vestibule. Furthermore, amiloride inhibition resulted in the channel occlusion in the middle of the pore. Summarizing, the ionic pore of FaNaC includes a large aqueous cavity, with a charge selectivity filter in the outer vestibule and the gate close to the interior.
Figures
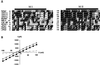



Similar articles
-
Electrostatic charge at position 552 affects the activation and permeation of FMRFamide-gated Na+ channels.J Physiol Sci. 2014 Mar;64(2):141-50. doi: 10.1007/s12576-013-0303-6. Epub 2014 Jan 12. J Physiol Sci. 2014. PMID: 24415456 Free PMC article.
-
Structural determinants of the closed KCa3.1 channel pore in relation to channel gating: results from a substituted cysteine accessibility analysis.J Gen Physiol. 2007 Apr;129(4):299-315. doi: 10.1085/jgp.200609726. Epub 2007 Mar 12. J Gen Physiol. 2007. PMID: 17353352 Free PMC article.
-
A tryptophan residue (W736) in the amino-terminus of the P-segment of domain II is involved in pore formation in Na(v)1.4 voltage-gated sodium channels.Pflugers Arch. 2002 Oct;445(1):18-24. doi: 10.1007/s00424-002-0887-9. Epub 2002 Aug 14. Pflugers Arch. 2002. PMID: 12397382
-
Insight toward epithelial Na+ channel mechanism revealed by the acid-sensing ion channel 1 structure.IUBMB Life. 2008 Sep;60(9):620-8. doi: 10.1002/iub.89. IUBMB Life. 2008. PMID: 18459164 Review.
-
Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure.Physiol Rev. 2002 Jul;82(3):735-67. doi: 10.1152/physrev.00007.2002. Physiol Rev. 2002. PMID: 12087134 Review.
Cited by
-
Intrinsic voltage dependence of the epithelial Na+ channel is masked by a conserved transmembrane domain tryptophan.J Biol Chem. 2009 Sep 18;284(38):25512-21. doi: 10.1074/jbc.M109.015917. Epub 2009 Jul 20. J Biol Chem. 2009. PMID: 19620245 Free PMC article.
-
A Drosophila DEG/ENaC subunit functions specifically in gustatory neurons required for male courtship behavior.J Neurosci. 2012 Mar 28;32(13):4665-74. doi: 10.1523/JNEUROSCI.6178-11.2012. J Neurosci. 2012. PMID: 22457513 Free PMC article.
-
Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.Nat Chem Biol. 2023 Oct;19(10):1276-1285. doi: 10.1038/s41589-023-01401-7. Epub 2023 Aug 7. Nat Chem Biol. 2023. PMID: 37550431
-
X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel.Cell. 2014 Feb 13;156(4):717-29. doi: 10.1016/j.cell.2014.01.011. Epub 2014 Feb 6. Cell. 2014. PMID: 24507937 Free PMC article.
-
The receptor site of the spider toxin PcTx1 on the proton-gated cation channel ASIC1a.J Physiol. 2006 Jan 15;570(Pt 2):339-54. doi: 10.1113/jphysiol.2005.095810. Epub 2005 Nov 10. J Physiol. 2006. PMID: 16284080 Free PMC article.
References
-
- Akabas M.H., Stauffer,D.A., Xu,M. and Karlin,A. (1992) Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science, 258, 307–310. - PubMed
-
- Askwith C.C., Cheng,C., Ikuma,M., Benson,C., Price,M.P. and Welsh,M.J. (2000) Neuropeptide FF and FMRF-amide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron, 26, 133–141. - PubMed
-
- Benson C.J., Eckert,S.P. and McCleskey,E.W. (1999) Acid-evoked currents in cardiac sensory neurons: A possible mediator of myocardial ischemic sensation. Circ. Res., 84, 921–928. - PubMed
-
- Canessa C.M., Merillat,A.M. and Rossier,B.C. (1994) Membrane topology of the epithelial sodium channel in intact cells. Am. J. Physiol., 267, C1682–C1690. - PubMed
-
- Chalfie M.and Wolinski,E. (1990) The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans. Nature, 345, 410–416. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

