Characterization of FasG segments required for 987P fimbria-mediated binding to piglet glycoprotein receptors
- PMID: 11598031
- PMCID: PMC100036
- DOI: 10.1128/IAI.69.11.6625-6632.2001
Characterization of FasG segments required for 987P fimbria-mediated binding to piglet glycoprotein receptors
Abstract
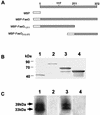
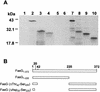
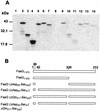
The 987P fimbriae of enterotoxigenic strains of Escherichia coli bind to both glycoprotein and glycolipid receptors on the brush borders of piglet enterocytes. A mutation in lysine residue 117 of the adhesive subunit FasG [fasG(K117A)] previously shown to abrogate 987P binding to the lipid receptor sulfatide did not affect the interaction with the glycoprotein receptors. Both the fimbriae and the FasG subunits of the wild type and the fasG(K117A) mutant bound to the glycoprotein receptors, confirming that lysine 117 was not required for binding to the glycoprotein receptors. Truncated FasG molecules were used to identify domains required for glycoprotein receptor recognition. At least two segments which did not include lysine117, namely, residues 211 (glutamine) to 220 (serine) and 20 (aspartic acid) to 41 (serine), were shown to be involved in the FasG-glycoprotein receptor interactions by ligand-blotting assays. Changing isoleucine 217 or leucine 215 of FasG to alanine abolished the property of a truncated FasG fusion protein to inhibit 987P recognition of its glycoprotein receptors. Thus, the K117 residue of FasG is required only for binding to the glycolipid receptor, whereas the newly identified hydrophobic residues of the FasG subunit are required specifically for the recognition of the glycoprotein receptor. Taken together, our data indicate that different residues of the FasG adhesin are important in 987P fimbrial binding to sulfatide and glycoprotein receptors, suggesting different mechanisms of interaction.
Figures
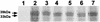

References
-
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

