Adaptins: the final recount
- PMID: 11598180
- PMCID: PMC60144
- DOI: 10.1091/mbc.12.10.2907
Adaptins: the final recount
Abstract
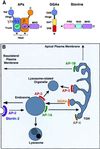
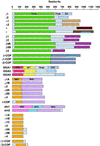
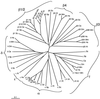
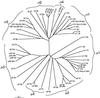
Adaptins are subunits of adaptor protein (AP) complexes involved in the formation of intracellular transport vesicles and in the selection of cargo for incorporation into the vesicles. In this article, we report the results of a survey for adaptins from sequenced genomes including those of man, mouse, the fruit fly Drosophila melanogaster, the nematode Caenorhabditis elegans, the plant Arabidopsis thaliana, and the yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe. We find that humans, mice, and Arabidopsis thaliana have four AP complexes (AP-1, AP-2, AP-3, and AP-4), whereas D. melanogaster, C. elegans, S. cerevisiae, and S. pombe have only three (AP-1, AP-2, and AP-3). Additional diversification of AP complexes arises from the existence of adaptin isoforms encoded by distinct genes or resulting from alternative splicing of mRNAs. We complete the assignment of adaptins to AP complexes and provide information on the chromosomal localization, exon-intron structure, and pseudogenes for the different adaptins. In addition, we discuss the structural and evolutionary relationships of the adaptins and the genetic analyses of their function. Finally, we extend our survey to adaptin-related proteins such as the GGAs and stonins, which contain domains homologous to the adaptins.
Figures


References
-
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Aguilar RC, Boehm M, Gorshkova I, Crouch RJ, Tomita K, Saito T, Ohno H, Bonifacino JS. Signal-binding specificity of the μ4 subunit of the adaptor protein complex AP-4. J Biol Chem. 2001;276:13145–13152. - PubMed
-
- Aguilar RC, Ohno H, Roche KW, Bonifacino JS. Functional domain mapping of the clathrin-associated adaptor medium chains μ1 and μ2. J Biol Chem. 1997;272:27160–27166. - PubMed
-
- Ball CL, Hunt SP, Robinson MS. Expression and localization of α-adaptin isoforms. J Cell Sci. 1995;108:2865–2875. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

