Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase?
- PMID: 11598227
- PMCID: PMC125088
Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase?
Abstract
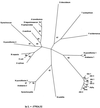
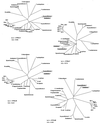
Although cellulose biosynthesis among the cyanobacteria has been suggested previously, we present the first conclusive evidence, to our knowledge, of the presence of cellulose in these organisms. Based on the results of x-ray diffraction, electron microscopy of microfibrils, and cellobiohydrolase I-gold labeling, we report the occurrence of cellulose biosynthesis in nine species representing three of the five sections of cyanobacteria. Sequence analysis of the genomes of four cyanobacteria revealed the presence of multiple amino acid sequences bearing the DDD35QXXRW motif conserved in all cellulose synthases. Pairwise alignments demonstrated that CesAs from plants were more similar to putative cellulose synthases from Anabaena sp. Pasteur Culture Collection 7120 and Nostoc punctiforme American Type Culture Collection 29133 than any other cellulose synthases in the database. Multiple alignments of putative cellulose synthases from Anabaena sp. Pasteur Culture Collection 7120 and N. punctiforme American Type Culture Collection 29133 with the cellulose synthases of other prokaryotes, Arabidopsis, Gossypium hirsutum, Populus alba x Populus tremula, corn (Zea mays), and Dictyostelium discoideum showed that cyanobacteria share an insertion between conserved regions U1 and U2 found previously only in eukaryotic sequences. Furthermore, phylogenetic analysis indicates that the cyanobacterial cellulose synthases share a common branch with CesAs of vascular plants in a manner similar to the relationship observed with cyanobacterial and chloroplast 16s rRNAs, implying endosymbiotic transfer of CesA from cyanobacteria to plants and an ancient origin for cellulose synthase in eukaryotes.
Figures
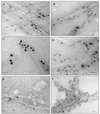


 ) have
reflections of 3.9
) have
reflections of 3.9  . For the −101 spacings, all genera with
the exception of Gloeocapsa (5.4
. For the −101 spacings, all genera with
the exception of Gloeocapsa (5.4  ) have 5.3-
) have 5.3- reflections. For the 101 spacings, all genera with the exception of
P. autumnale (5.9
reflections. For the 101 spacings, all genera with the exception of
P. autumnale (5.9  ) have 6.0-
) have 6.0- reflections.
The presence of contaminating crystalline materials is evidenced by the
existence of peaks not related to cellulose. Note that these peaks do
not always produce uniform overlaps with all four
genera.
reflections.
The presence of contaminating crystalline materials is evidenced by the
existence of peaks not related to cellulose. Note that these peaks do
not always produce uniform overlaps with all four
genera.

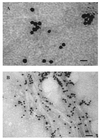
Similar articles
-
Beta-D-glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1-->3),(1-->4)beta-D-glucan synthase.Plant Mol Biol. 2001 Sep;47(1-2):145-60. Plant Mol Biol. 2001. PMID: 11554469
-
Cellulose synthase (CesA) genes in the green alga Mesotaenium caldariorum.Eukaryot Cell. 2002 Dec;1(6):847-55. doi: 10.1128/EC.1.6.847-855.2002. Eukaryot Cell. 2002. PMID: 12477785 Free PMC article.
-
Xylem-specific and tension stress-responsive expression of cellulose synthase genes from aspen trees.Appl Biochem Biotechnol. 2003 Spring;105 -108:17-25. doi: 10.1385/abab:105:1-3:17. Appl Biochem Biotechnol. 2003. PMID: 12721472
-
Higher plant cellulose synthases.Genome Biol. 2000;1(4):REVIEWS3001. doi: 10.1186/gb-2000-1-4-reviews3001. Epub 2000 Oct 13. Genome Biol. 2000. PMID: 11178255 Free PMC article. Review.
-
Cellulose synthases and synthesis in Arabidopsis.Mol Plant. 2011 Mar;4(2):199-211. doi: 10.1093/mp/ssq079. Epub 2011 Feb 9. Mol Plant. 2011. PMID: 21307367 Review.
Cited by
-
Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida.Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):5247-52. doi: 10.1073/pnas.1221259110. Epub 2013 Mar 15. Proc Natl Acad Sci U S A. 2013. PMID: 23503846 Free PMC article.
-
Complex organic matter degradation by secondary consumers in chemolithoautotrophy-based subsurface geothermal ecosystems.PLoS One. 2023 Aug 18;18(8):e0281277. doi: 10.1371/journal.pone.0281277. eCollection 2023. PLoS One. 2023. PMID: 37594978 Free PMC article.
-
Cyclic di-GMP: the first 25 years of a universal bacterial second messenger.Microbiol Mol Biol Rev. 2013 Mar;77(1):1-52. doi: 10.1128/MMBR.00043-12. Microbiol Mol Biol Rev. 2013. PMID: 23471616 Free PMC article. Review.
-
CugP is a novel ubiquitous non-GalU-type bacterial UDP-glucose pyrophosphorylase found in cyanobacteria.J Bacteriol. 2014 Jul;196(13):2348-54. doi: 10.1128/JB.01591-14. Epub 2014 Apr 11. J Bacteriol. 2014. PMID: 24727225 Free PMC article.
-
Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria.Front Microbiol. 2015 Nov 17;6:1282. doi: 10.3389/fmicb.2015.01282. eCollection 2015. Front Microbiol. 2015. PMID: 26635751 Free PMC article. Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barclay W, Lewin R. Microalgal polysaccharide production for the conditioning of agricultural soils. Plant and Soil. 1985;88:159–169.
-
- Brown RM., Jr Cellulose microfibril assembly and orientation: recent developments. J Cell Sci Suppl. 1985;2:13–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
