Arabidopsis thaliana type I and II chaperonins
- PMID: 11599560
- PMCID: PMC434400
- DOI: 10.1379/1466-1268(2001)006<0190:attiai>2.0.co;2
Arabidopsis thaliana type I and II chaperonins
Abstract
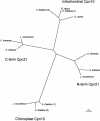
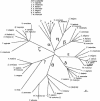
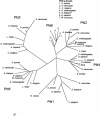
An examination of the Arabidopsis thaliana genome sequence led to the identification of 29 predicted genes with the potential to encode members of the chaperonin family of chaperones (CPN60 and CCT), their associated cochaperonins, and the cytoplasmic chaperonin cofactor prefoldin. These comprise the first complete set of plant chaperonin protein sequences and indicate that the CPN family is more diverse than previously described. In addition to surprising sequence diversity within CPN subclasses, the genomic data also suggest the existence of previously undescribed family members, including a 10-kDa chloroplast cochaperonin. Consideration of the sequence data described in this review prompts questions about the complexities of plant CPN systems and the evolutionary relationships and functions of the component proteins, most of which have not been studied experimentally.
Figures


References
-
- Ahnert V, May C, Gerke R, Kindl H. Cucumber T-complex protein: molecular cloning, bacterial expression and characterization within a 22-S cytosolic complex in cotyledons and hypocotyls. Eur J Biochem. 1996;235:114–119. - PubMed
-
- Archibald JM, Logsdon JM, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol. 2000;17:1456–1466. - PubMed
-
- Baneyx F, Bertsch U, Kalbach CE, van der Vies SM, Soll J, Gatenby AA. Spinach chloroplast cpn21 co-chaperonin possesses two functional domains fused together in a toroidal structure and exhibits nucleotide-dependent binding to plastid chaperonin 60. J Biol Chem. 1995;270:10695–10702. - PubMed
-
- Bertsch U, Soll J. Functional analysis of isolated cpn10 domains and conserved amino acid residues in spinach chloroplast co-chaperonin by site-directed mutagenesis. Plant Mol Biol. 1995;29:1039–1055. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
