Arabidopsis thaliana Hsp100 proteins: kith and kin
- PMID: 11599563
- PMCID: PMC434403
- DOI: 10.1379/1466-1268(2001)006<0219:athpka>2.0.co;2
Arabidopsis thaliana Hsp100 proteins: kith and kin
Abstract
Arabidopsis thaliana, the first plant for which the entire genome sequence is available, was also among the first plant species from which Hsp100 proteins were characterized. The Athsp101 complementary DNA (cDNA) corresponds to the gene identification At1g74310 in the Arabidopsis genome sequence. Analysis of the genome revealed 7 additional proteins that are variably homologous with At1g74310 throughout the entire amino acid sequence and significant similarities or identities in the signature sequences conserved among Hsp100 proteins. Although AtHsp101 is cytoplasmic, 5 of the 7 related proteins have predicted plastidial localization signals. This complete description of the AtHsp100 family sets the stage for future research on expression and function.
Figures


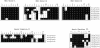
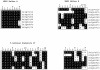
References
-
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. - PubMed
-
- Hong SW, Vierling E 2001 Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J in press. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
