A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences
- PMID: 11600698
- PMCID: PMC60209
- DOI: 10.1093/nar/29.20.4097
A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences
Abstract
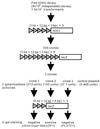
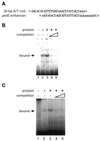
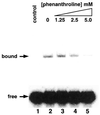
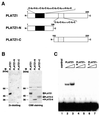
Complementary DNA encoding a DNA-binding protein, designated PLATZ1 (plant AT-rich sequence- and zinc-binding protein 1), was isolated from peas. The amino acid sequence of the protein is similar to those of other uncharacterized proteins predicted from the genome sequences of higher plants. However, no paralogous sequences have been found outside the plant kingdom. Multiple alignments among these paralogous proteins show that several cysteine and histidine residues are invariant, suggesting that these proteins are a novel class of zinc-dependent DNA-binding proteins with two distantly located regions, C-x(2)-H-x(11)-C-x(2)-C-x((4-5))-C-x(2)-C-x((3-7))-H-x(2)-H and C-x(2)-C-x((10-11))-C-x(3)-C. In an electrophoretic mobility shift assay, the zinc chelator 1,10-o-phenanthroline inhibited DNA binding, and two distant zinc-binding regions were required for DNA binding. A protein blot with (65)ZnCl(2) showed that both regions are required for zinc-binding activity. The PLATZ1 protein non-specifically binds to A/T-rich sequences, including the upstream region of the pea GTPase pra2 and plastocyanin petE genes. Expression of the PLATZ1 repressed those of the reporter constructs containing the coding sequence of luciferase gene driven by the cauliflower mosaic virus (CaMV) 35S90 promoter fused to the tandem repeat of the A/T-rich sequences. These results indicate that PLATZ1 is a novel class of plant-specific zinc-dependent DNA-binding protein responsible for A/T-rich sequence-mediated transcriptional repression.
Figures




References
-
- Riechmann J.L., Heard,J., Martin,G., Reuber,L., Jiang,C., Keddie,J., Adam,L., Pineda,O., Ratcliffe,O.J., Samaha,R.R., Creelman,R., Pilgrim,M., Broun,P., Zhang,J.Z., Ghandehari,D., Sherman,B.K. and Yu,G. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 290, 2105–2110. - PubMed
-
- Inaba T., Nagano,Y., Reid,J.B. and Sasaki,Y. (2000) DE1, a 12-base pair cis-regulatory element sufficient to confer dark-inducible and light down-regulated expression to a minimal promoter in pea. J. Biol. Chem., 275, 19723–19727. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

