Pyret, a Ty3/Gypsy retrotransposon in Magnaporthe grisea contains an extra domain between the nucleocapsid and protease domains
- PMID: 11600699
- PMCID: PMC60222
- DOI: 10.1093/nar/29.20.4106
Pyret, a Ty3/Gypsy retrotransposon in Magnaporthe grisea contains an extra domain between the nucleocapsid and protease domains
Abstract
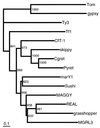
A novel Ty3/Gypsy retrotransposon, named Pyret, was identified in the plant pathogenic fungus Magnaporthe grisea (anamorph Pyricularia oryzae). Pyret-related elements were distributed in a wide range of Pyricularia isolates from various gramineous plants. The Pyret element is 7250 bp in length with a 475 bp LTR and one conceptual ORF. The ORF contains seven nonsense mutations in the reading frame, indicating that the Pyret clone is lightly degenerate. Comparative domain analysis among retroelements revealed that Pyret exhibits an extra domain (WCCH domain) beyond the basic components of LTR retrotransposons. The WCCH domain consists of approximately 300 amino acids and is located downstream of the nucleocapsid domain. The WCCH domain is so named because it contains two repeats of a characteristic amino acid sequence, W-X(2)-C-X(4)-C-X(2)-H-X(3)-K. A WCCH motif-like sequence is found in the precoat protein of some geminiviruses, viral RNA-dependent RNA polymerase and also in an Arabidopsis protein of unknown function. Interestingly, detailed sequence analysis of the gag protein revealed that Pyret, as well as some other chromodomain-containing LTR retrotransposons, displays significant sequence homology with members of the gammaretroviruses (MLV-related retroviruses) in the capsid and nucleocapsid domains. This suggests that chromodomain-containing LTR retrotransposons and gammaretroviruses may share a common ancestor with the gag protein.
Figures
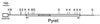

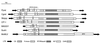


Similar articles
-
REAL, an LTR retrotransposon from the plant pathogenic fungus Alternaria alternata.Mol Gen Genet. 2000 May;263(4):625-34. doi: 10.1007/s004380051210. Mol Gen Genet. 2000. PMID: 10852484
-
Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis.BMC Evol Biol. 2008 Oct 8;8:276. doi: 10.1186/1471-2148-8-276. BMC Evol Biol. 2008. PMID: 18842133 Free PMC article.
-
Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins.Genetics. 1998 Jun;149(2):703-15. doi: 10.1093/genetics/149.2.703. Genetics. 1998. PMID: 9611185 Free PMC article.
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
-
Function of a retrotransposon nucleocapsid protein.RNA Biol. 2010 Nov-Dec;7(6):642-54. doi: 10.4161/rna.7.6.14117. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21189452 Free PMC article. Review.
Cited by
-
The telomere-linked helicase (TLH) gene family in Magnaporthe oryzae: revised gene structure reveals a novel TLH-specific protein motif.Curr Genet. 2009 Jun;55(3):253-62. doi: 10.1007/s00294-009-0240-3. Epub 2009 Apr 10. Curr Genet. 2009. PMID: 19360408
-
Characterization of AFLAV, a Tf1/Sushi retrotransposon from Aspergillus flavus.Mycopathologia. 2007 Feb;163(2):97-104. doi: 10.1007/s11046-006-0088-8. Epub 2007 Feb 7. Mycopathologia. 2007. PMID: 17286166
-
Occan, a novel transposon in the Fot1 family, is ubiquitously found in several Magnaporthe grisea isolates.Curr Genet. 2003 Mar;42(6):322-31. doi: 10.1007/s00294-002-0365-0. Epub 2003 Feb 8. Curr Genet. 2003. PMID: 12612805
-
Genome-Wide Comparison of Magnaporthe Species Reveals a Host-Specific Pattern of Secretory Proteins and Transposable Elements.PLoS One. 2016 Sep 22;11(9):e0162458. doi: 10.1371/journal.pone.0162458. eCollection 2016. PLoS One. 2016. PMID: 27658241 Free PMC article.
-
Characterization of chromosome ends in the filamentous fungus Neurospora crassa.Genetics. 2009 Mar;181(3):1129-45. doi: 10.1534/genetics.107.084392. Epub 2008 Dec 22. Genetics. 2009. PMID: 19104079 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

