Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly
- PMID: 11600704
- PMCID: PMC60220
- DOI: 10.1093/nar/29.20.4154
Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly
Abstract
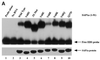
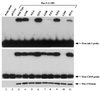
Combinatorial association of DNA-binding proteins on composite binding sites enhances their nucleotide sequence specificity and functional synergy. As a paradigm for these interactions, Pax-5 (BSAP) assembles ternary complexes with Ets proteins on the B cell-specific mb-1 promoter through interactions between their respective DNA-binding domains. Pax-5 recruits Ets-1 to bind the promoter, but not the closely related Ets protein SAP1a. Here we show that, while several different mutations increase binding of SAP1a to an optimized Ets binding site, only conversion of Val68 to an acidic amino acid facilitates ternary complex assembly with Pax-5 on the mb-1 promoter. This suggests that enhanced DNA binding by SAP1a is not sufficient for recruitment by Pax-5, but instead involves protein-protein interactions mediated by the acidic side chain. Recruitment of Ets proteins by Pax-5 requires Gln22 within the N-terminal beta-hairpin motif of its paired domain. The beta-hairpin also participates in recognition of a subset of Pax-5-binding sites. Thus, Pax-5 incorporates protein-protein interaction and DNA recognition functions in a single motif. The Caenorhabditis elegans Pax protein EGL-38 also binds specifically to the mb-1 promoter and recruits murine Ets-1 or the C.elegans Ets protein T08H4.3, but not the related LIN-1 protein. Together, our results define specific amino acid requirements for Pax-Ets ternary complex assembly and show that the mechanism is conserved between evolutionarily related proteins of diverse animal species. Moreover, the data suggest that interactions between Pax and Ets proteins are an important mechanism that regulates fundamental biological processes in worms and humans.
Figures










Similar articles
-
Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP).Nucleic Acids Res. 2003 Oct 1;31(19):5483-9. doi: 10.1093/nar/gkg785. Nucleic Acids Res. 2003. PMID: 14500810 Free PMC article.
-
Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter.Genes Dev. 1996 Sep 1;10(17):2198-211. doi: 10.1101/gad.10.17.2198. Genes Dev. 1996. PMID: 8804314
-
The highly conserved beta-hairpin of the paired DNA-binding domain is required for assembly of Pax-Ets ternary complexes.Mol Cell Biol. 1999 Mar;19(3):2231-41. doi: 10.1128/MCB.19.3.2231. Mol Cell Biol. 1999. PMID: 10022910 Free PMC article.
-
Regulation of Ets function by protein - protein interactions.Oncogene. 2000 Dec 18;19(55):6514-23. doi: 10.1038/sj.onc.1204035. Oncogene. 2000. PMID: 11175367 Review.
-
Ets-1 flips for new partner Pax-5.Structure. 2002 Jan;10(1):11-4. doi: 10.1016/s0969-2126(01)00701-8. Structure. 2002. PMID: 11796106 Review.
Cited by
-
Genomic and biochemical insights into the specificity of ETS transcription factors.Annu Rev Biochem. 2011;80:437-71. doi: 10.1146/annurev.biochem.79.081507.103945. Annu Rev Biochem. 2011. PMID: 21548782 Free PMC article. Review.
-
Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP).Nucleic Acids Res. 2003 Oct 1;31(19):5483-9. doi: 10.1093/nar/gkg785. Nucleic Acids Res. 2003. PMID: 14500810 Free PMC article.
-
Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter.Mol Cell Biol. 2002 Dec;22(24):8539-51. doi: 10.1128/MCB.22.24.8539-8551.2002. Mol Cell Biol. 2002. PMID: 12446773 Free PMC article.
-
Highly cooperative recruitment of Ets-1 and release of autoinhibition by Pax5.J Mol Biol. 2009 Sep 18;392(2):452-64. doi: 10.1016/j.jmb.2009.07.028. Epub 2009 Jul 17. J Mol Biol. 2009. PMID: 19616560 Free PMC article.
-
De Novo Mutations in EBF3 Cause a Neurodevelopmental Syndrome.Am J Hum Genet. 2017 Jan 5;100(1):138-150. doi: 10.1016/j.ajhg.2016.11.020. Epub 2016 Dec 23. Am J Hum Genet. 2017. PMID: 28017370 Free PMC article.
References
-
- Czerny T., Schaffner,G. and Busslinger,M. (1993) DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev., 7, 2048–2061. - PubMed
-
- Xu W., Rould,M.A., Jun,S., Desplan,C. and Pabo,C.O. (1995) Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell, 80, 639–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

