A novel yeast system for in vivo selection of recognition sequences: defining an optimal c-Myb-responsive element
- PMID: 11600718
- PMCID: PMC60227
- DOI: 10.1093/nar/29.20.e99
A novel yeast system for in vivo selection of recognition sequences: defining an optimal c-Myb-responsive element
Abstract
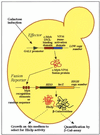
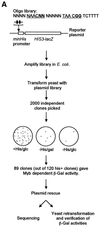
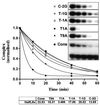
Yeast (Saccharomyces cerevisiae) has proved to be a highly valuable tool in a range of screening methods. We present in this work the design and use of a novel yeast effector-reporter system for selection of sequences recognised by DNA-binding proteins in vivo. A dual HIS3-lacZ reporter under the control of a single randomised response element facilitates both positive growth selection of binding sequences and subsequent quantification of the strength of the selected sequence. A galactose-inducible effector allows discrimination between reporter activation caused by the protein under study and activation due to endogenous factors. The system mimics the physiological gene dosage relationship between transcription factor and target genes in vivo by using a low copy effector plasmid and a high copy reporter plasmid, favouring sequence selectivity. The utility of the novel yeast screening system was demonstrated by using it to refine the definition of an optimal recognition element for the c-Myb transcription factor (MRE). We present screening data supporting an extended MRE consensus closely mimicking known strong response elements and where a sequence of 11 nt influences activity. Novel features include a more strict sequence requirement in the second half-site of the MRE where a T-rich sequence is preferred in vivo.
Figures








References
-
- Brent R. and Finley,R.L.Jr (1997) Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet., 31, 663––704.. - PubMed
-
- Fields S. and Song,O. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245––246.. - PubMed
-
- Drees B.L. (1999) Progress and variations in two-hybrid and three-hybrid technologies. Curr. Opin. Chem. Biol., 3, 64––70.. - PubMed
-
- Mendelsohn A.R. and Brent,R. (1999) Protein interaction methods—toward an endgame. Science, 284, 1948––1950.. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

