Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase
- PMID: 11602641
- PMCID: PMC2193509
- DOI: 10.1084/jem.194.8.1123
Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase
Abstract
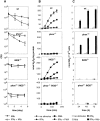
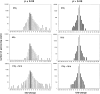
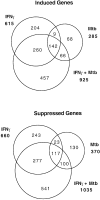
Macrophage activation determines the outcome of infection by Mycobacterium tuberculosis (Mtb). Interferon-gamma (IFN-gamma) activates macrophages by driving Janus tyrosine kinase (JAK)/signal transducer and activator of transcription-dependent induction of transcription and PKR-dependent suppression of translation. Microarray-based experiments reported here enlarge this picture. Exposure to IFN-gamma and/or Mtb led to altered expression of 25% of the monitored genome in macrophages. The number of genes suppressed by IFN-gamma exceeded the number of genes induced, and much of the suppression was transcriptional. Five times as many genes related to immunity and inflammation were induced than suppressed. Mtb mimicked or synergized with IFN-gamma more than antagonized its actions. Phagocytosis of nonviable Mtb or polystyrene beads affected many genes, but the transcriptional signature of macrophages infected with viable Mtb was distinct. Studies involving macrophages deficient in inducible nitric oxide synthase and/or phagocyte oxidase revealed that these two antimicrobial enzymes help orchestrate the profound transcriptional remodeling that underlies macrophage activation.
Figures
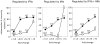


References
-
- Schreiber R.D., Pace J.L., Russell S.W., Altman A., Katz D.H. Macrophage-activating factor produced by a T cell hybridomaphysiochemical and biosynthetic resemblance to γ-interferon. J. Immunol. 1983;131:826–832. - PubMed
-
- Jouanguy E., Lamhamedi-Cherradi S., Lammas D., Dorman S.E., Fondaneche M.C., Dupuis S., Doffinger R., Altare F., Girdlestone J., Emile J.F. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 1999;21:370–378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

