Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA
- PMID: 11602702
- PMCID: PMC114642
- DOI: 10.1128/JVI.75.22.10603-10611.2001
Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA
Abstract
oriP is a 1.7-kb region of the Epstein-Barr virus (EBV) chromosome that supports replication and stable maintenance of plasmids in human cells that contain EBV-encoded protein EBNA1. Plasmids that depend on oriP are replicated once per cell cycle by cellular factors. The replicator of oriP is an approximately 120-bp region called DS which depends on either of two pairs of closely spaced EBNA1 binding sites. Here we report that changing the distance between the EBNA1 sites of a functional pair by inserting or deleting 1 or 2 bp abolished replication activity. The results indicated that, while the distance separating the binding sites is critical, the specific nucleotide sequence between them is unlikely to be important. The use of electrophoretic mobility shift assays to investigate binding by EBNA1 to the sites with normal or altered spacing revealed that EBNA1 induces DNA to bend significantly when it binds, with the center of bending coinciding with the center of binding. EBNA1 binding to a functional pair of sites which are spaced 21 bp apart center to center and which thus are in helical phase induces a larger symmetrical bend, which based on electrophoretic mobility approximates the sum of two separate EBNA1-induced DNA bends. The results imply that replication from oriP requires a precise structure in which DNA forms a large bend around two EBNA1 dimers.
Figures


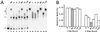
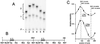
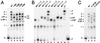
References
-
- Blow J J, Laskey R A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. - PubMed
-
- Bochkarev A, Barwell J A, Pfuetzner R A, Bochkareva E, Frappier L, Edwards A M. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

