Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41
- PMID: 11602729
- PMCID: PMC114669
- DOI: 10.1128/JVI.75.22.10892-10905.2001
Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41
Abstract
The identification and epitope mapping of broadly neutralizing anti-human immunodeficiency virus type 1 (HIV-1) antibodies (Abs) is important for vaccine design, but, despite much effort, very few such Abs have been forthcoming. Only one broadly neutralizing anti-gp41 monoclonal Ab (MAb), 2F5, has been described. Here we report on two MAbs that recognize a region immediately C-terminal of the 2F5 epitope. Both MAbs were generated from HIV-1-seropositive donors, one (Z13) from an antibody phage display library, and one (4E10) as a hybridoma. Both MAbs recognize a predominantly linear and relatively conserved epitope, compete with each other for binding to synthetic peptide derived from gp41, and bind to HIV-1(MN) virions. By flow cytometry, these MAbs appear to bind relatively weakly to infected cells and this binding is not perturbed by pretreatment of the infected cells with soluble CD4. Despite the apparent linear nature of the epitopes of Z13 and 4E10, denaturation of recombinant envelope protein reduces the binding of these MAbs, suggesting some conformational requirements for full epitope expression. Most significantly, Z13 and 4E10 are able to neutralize selected primary isolates from diverse subtypes of HIV-1 (e.g., subtypes B, C, and E). The results suggest that a rather extensive region of gp41 close to the transmembrane domain is accessible to neutralizing Abs and could form a useful target for vaccine design.
Figures



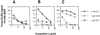





References
-
- Baba T W, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini L A, Posner M R, Katinger H, Stiegler G, Bernacky B J, Rizvi T A, Schmidt R, Hill L R, Keeling M E, Lu Y, Wright J E, Chou T C, Ruprecht R M. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. - PubMed
-
- Barbas C F, III, Burton D R, Scott J K, Silverman G J. Phage display: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001.
-
- Barbas C F, III, Collet T A, Amberg W, Roben P, Binley J M, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, Haigwood N L, Cabezas E, Satterthwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. - PubMed
-
- Binley J M, Ditzel H J, Barbas III C F, Sullivan N, Sodroski J, Parren P W H I, Burton D R. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retrovir. 1996;12:911–924. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- GM46192/GM/NIGMS NIH HHS/United States
- AI39420/AI/NIAID NIH HHS/United States
- R37 AI036082/AI/NIAID NIH HHS/United States
- M01 RR00833/RR/NCRR NIH HHS/United States
- R37 AI033292/AI/NIAID NIH HHS/United States
- R01 AI033292/AI/NIAID NIH HHS/United States
- AI36082/AI/NIAID NIH HHS/United States
- AI44293/AI/NIAID NIH HHS/United States
- AI33292/AI/NIAID NIH HHS/United States
- R01 GM046192/GM/NIGMS NIH HHS/United States
- M01 RR000833/RR/NCRR NIH HHS/United States
- R01 AI039420/AI/NIAID NIH HHS/United States
- AI40377/AI/NIAID NIH HHS/United States
- HL59735/HL/NHLBI NIH HHS/United States
- R01 AI036082/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

