Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus
- PMID: 11602766
- PMCID: PMC114706
- DOI: 10.1128/JVI.75.22.11244-11248.2001
Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus
Abstract
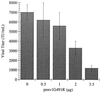
Mice expressing the Fv-4 gene are resistant to infection by ecotropic murine leukemia viruses (MuLVs). The Fv-4 gene encodes an envelope (Env) protein whose putative receptor-binding domain resembles that of ecotropic MuLV Env protein. Resistance to ecotropic MuLVs appears to result from viral interference involving binding of the endogenously expressed Fv-4 env-encoded protein to the ecotropic receptor, although the immune system also plays a role in resistance. The Fv-4 env-encoded protein is processed normally and can be incorporated into virus particles but is unable to promote viral entry. Among the many sequence variations between the transmembrane (TM) subunit of the Fv-4 env-encoded protein and the TM subunits of other MuLV Env proteins, there is a substitution of an arginine residue in the Fv-4 env-encoded protein for a glycine residue (gly-491 in Moloney MuLV Env) that is otherwise conserved in all of the other MuLVs. This residue is present in the MuLV TM fusion peptide sequence. In this study, gly-491 of Moloney MuLV Env has been replaced with other residues and a mutant Env bearing a substitution for gly-487 was also created. G491R recapitulates the Fv-4 Env phenotype in cell culture, indicating that this substitution is sufficient for creation of an Env protein that can establish the interference-mediated resistance to ecotropic viruses produced by the Fv-4 gene. Analysis of the mutant MuLV Env proteins also has implications for an understanding of the role of conserved glycine residues in fusion peptides and for the engineering of organismal resistance to retroviruses.
Figures

References
-
- Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. - PubMed
-
- Bonnafous P, Stegmann T. Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J Biol Chem. 2000;275:6160–6166. - PubMed
-
- Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. - PubMed
-
- Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science. 1997;277:1662–1666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

