RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization
- PMID: 11604510
- PMCID: PMC99945
- DOI: 10.1128/MCB.21.22.7747-7760.2001
RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization
Abstract
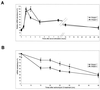
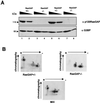
Mitogen activation of mRNA decay pathways likely involves specific endoribonucleases, such as G3BP, a phosphorylation-dependent endoribonuclease that associates with RasGAP in dividing but not quiescent cells. G3BP exclusively cleaves between cytosine and adenine (CA) after a specific interaction with RNA through the carboxyl-terminal RRM-type RNA binding motif. Accordingly, G3BP is tightly associated with a subset of poly(A)(+) mRNAs containing its high-affinity binding sequence, such as the c-myc mRNA in mouse embryonic fibroblasts. Interestingly, c-myc mRNA decay is delayed in RasGAP-deficient fibroblasts, which contain a defective isoform of G3BP that is not phosphorylated at serine 149. A G3BP mutant in which this serine is changed to alanine remains exclusively cytoplasmic, whereas a glutamate for serine substitution that mimics the charge of a phosphorylated serine is translocated to the nucleus. Thus, a growth factor-induced change in mRNA decay may be modulated by the nuclear localization of a site-specific endoribonuclease such as G3BP.
Figures





References
-
- Belasco J, Brawerman G. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993.
-
- Bernstein P, Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989;14:373–377. - PubMed
-
- Bernstein P L, Herrick D J, Prokipcak R D, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992;6:642–654. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
