N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo
- PMID: 11604512
- PMCID: PMC99947
- DOI: 10.1128/MCB.21.22.7775-7786.2001
N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo
Abstract
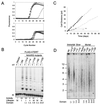
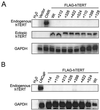
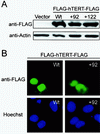
Most tumor cells depend upon activation of the ribonucleoprotein enzyme telomerase for telomere maintenance and continual proliferation. The catalytic activity of this enzyme can be reconstituted in vitro with the RNA (hTR) and catalytic (hTERT) subunits. However, catalytic activity alone is insufficient for the full in vivo function of the enzyme. In addition, the enzyme must localize to the nucleus, recognize chromosome ends, and orchestrate telomere elongation in a highly regulated fashion. To identify domains of hTERT involved in these biological functions, we introduced a panel of 90 N-terminal hTERT substitution mutants into telomerase-negative cells and assayed the resulting cells for catalytic activity and, as a marker of in vivo function, for cellular proliferation. We found four domains to be essential for in vitro and in vivo enzyme activity, two of which were required for hTR binding. These domains map to regions defined by sequence alignments and mutational analysis in yeast, indicating that the N terminus has also been functionally conserved throughout evolution. Additionally, we discovered a novel domain, DAT, that "dissociates activities of telomerase," where mutations left the enzyme catalytically active, but was unable to function in vivo. Since mutations in this domain had no measurable effect on hTERT homomultimerization, hTR binding, or nuclear targeting, we propose that this domain is involved in other aspects of in vivo telomere elongation. The discovery of these domains provides the first step in dissecting the biological functions of human telomerase, with the ultimate goal of targeting this enzyme for the treatment of human cancers.
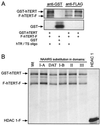
Figures


References
-
- Avilion A A, Piatyszek M A, Gupta J, Shay J W, Bacchetti S, Greider C W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. - PubMed
-
- Bacchetti S, Counter C M. Telomeres and telomerase in human cancer. Int J Oncol. 1995;7:423–432. - PubMed
-
- Bachand F, Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J Biol Chem. 1999;274:38027–38031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
