A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration
- PMID: 11604518
- PMCID: PMC99953
- DOI: 10.1128/MCB.21.22.7839-7851.2001
A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration
Abstract
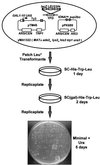
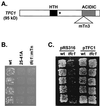
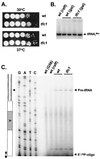
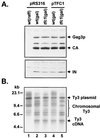
Position-specific integration of the retroviruslike element Ty3 near the transcription initiation sites of tRNA genes requires transcription factors IIIB and IIIC (TFIIIB and TFIIIC). Using a genetic screen, we isolated a mutant with a truncated 95-kDa subunit of TFIIIC (TFIIIC95) that reduced the apparent retrotransposition of Ty3 into a plasmid-borne target site between two divergently transcribed tRNA genes. Although TFIIIC95 is conserved and essential, no defect in growth or transcription of tRNAs was detected in the mutant. Steps of the Ty3 life cycle, such as protein expression, proteolytic processing, viruslike particle formation, and reverse transcription, were not affected by the mutation. However, Ty3 integration into a divergent tDNA target occurred exclusively in one orientation in the mutant strain. Investigation of this orientation bias showed that TFIIIC95 and Ty3 integrase interacted in two-hybrid and glutathione S-transferase pulldown assays and that interaction with the mutant TFIIIC95 protein was attenuated. The orientation bias observed here suggests that even for wild-type Ty3, the protein complexes associated with the long terminal repeats are not equivalent in vivo.
Figures


References
-
- Andrake M D, Skalka A M. Retroviral Integrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. - PubMed
-
- Arciszewska L K, Drake D, Craig N L. Transposon Tn7. cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. 4, section 20.1. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1999.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
