A hybrid bacterial replication origin
- PMID: 11606418
- PMCID: PMC1084124
- DOI: 10.1093/embo-reports/kve225
A hybrid bacterial replication origin
Abstract
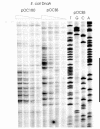
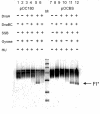
We constructed a hybrid replication origin that consists of the main part of oriC from Escherichia coli, the DnaA box region and the AT-rich region from Bacillus subtilis oriC. The AT-rich region could be unwound by E. coli DnaA protein, and the DnaB helicase was loaded into the single-stranded bubble. The results show that species specificity, i.e. which DnaA protein can do the unwinding, resides within the DnaA box region of oriC.
Figures

Similar articles
-
SSB promotes DnaB helicase passage through DnaA complexes at the replication origin oriC for bidirectional replication.J Biochem. 2025 Apr 2;177(4):305-316. doi: 10.1093/jb/mvaf003. J Biochem. 2025. PMID: 39776183 Free PMC article.
-
DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins.Gene. 1999 Mar 4;228(1-2):123-32. doi: 10.1016/s0378-1119(99)00009-8. Gene. 1999. PMID: 10072765
-
Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin.J Biol Chem. 2001 Nov 30;276(48):44919-25. doi: 10.1074/jbc.M107463200. Epub 2001 Sep 10. J Biol Chem. 2001. PMID: 11551962
-
Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading.Biochimie. 2001 Jan;83(1):5-12. doi: 10.1016/s0300-9084(00)01216-5. Biochimie. 2001. PMID: 11254968 Review.
-
DnaA structure, function, and dynamics in the initiation at the chromosomal origin.Plasmid. 2009 Sep;62(2):71-82. doi: 10.1016/j.plasmid.2009.06.003. Epub 2009 Jun 13. Plasmid. 2009. PMID: 19527752 Review.
Cited by
-
In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein.PLoS One. 2009;4(1):e4139. doi: 10.1371/journal.pone.0004139. Epub 2009 Jan 7. PLoS One. 2009. PMID: 19127296 Free PMC article.
-
Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria.Biochem J. 2005 Jul 15;389(Pt 2):471-81. doi: 10.1042/BJ20050143. Biochem J. 2005. PMID: 15790315 Free PMC article.
References
-
- Baker T.A., Funnell, B.E. and Kornberg, A. (1987) Helicase action of DnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J. Biol. Chem., 262, 6877–6885. - PubMed
-
- Bramhill D. and Kornberg, A. (1988) Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell, 52, 743–755. - PubMed
-
- Buhk H.J. and Messer, W. (1983) Replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene, 24, 265–279. - PubMed
-
- Echols H. (1990) Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J. Biol. Chem., 265, 14697–14700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

