T7 phage display: a novel genetic selection system for cloning RNA-binding proteins from cDNA libraries
- PMID: 11606722
- PMCID: PMC60806
- DOI: 10.1073/pnas.211439598
T7 phage display: a novel genetic selection system for cloning RNA-binding proteins from cDNA libraries
Abstract
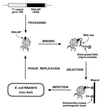
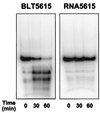
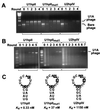
RNA-binding proteins are central to posttranscriptional gene regulation and play an important role in a number of major human diseases. Cloning such proteins is a crucial but often difficult step in elucidating the biological function of RNA regulatory elements. To make it easier to clone proteins that specifically bind RNA elements of interest, we have developed a rapid and broadly applicable in vitro genetic selection method based on T7 phage display. Using hairpin II of U1 small nuclear RNA (U1hpII) or the 3' stem loop of histone mRNA as bait, we could selectively amplify T7 phage that display either the spliceosomal protein U1A or the histone stem loop-binding protein from a lung cDNA phage library containing more than 10(7) independent clones. The use of U1hpII mutants with various affinities for U1A revealed that this method allows the selection even of proteins that bind their cognate RNA targets with relatively weak affinities (K(d) as high as the micromolar range). Experiments with a mixture of recombinant phage displaying U1A or the closely related protein U2B" demonstrated that addition of a competitor RNA can suppress selection of a protein with a higher affinity for a given RNA target, thereby allowing the preferential amplification of a lower affinity protein. Together, these findings suggest that T7 phage display can be used to rapidly and selectively clone virtually any protein that binds a known RNA regulatory element, including those that bind with low affinity or that must compete for binding with other proteins.
Figures


Similar articles
-
Phage display cDNA cloning of protein with carbohydrate affinity.Biochem Biophys Res Commun. 1999 Feb 16;255(2):194-9. doi: 10.1006/bbrc.1999.0175. Biochem Biophys Res Commun. 1999. PMID: 10049685
-
The role of RNA structure in the interaction of U1A protein with U1 hairpin II RNA.RNA. 2006 Jul;12(7):1168-78. doi: 10.1261/rna.75206. Epub 2006 May 31. RNA. 2006. PMID: 16738410 Free PMC article.
-
Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target.Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11859-63. doi: 10.1073/pnas.92.25.11859. Proc Natl Acad Sci U S A. 1995. PMID: 8524863 Free PMC article.
-
Advances in the T7 phage display system (Review).Mol Med Rep. 2018 Jan;17(1):714-720. doi: 10.3892/mmr.2017.7994. Epub 2017 Nov 7. Mol Med Rep. 2018. PMID: 29115536 Review.
-
T7 Phage as an Emerging Nanobiomaterial with Genetically Tunable Target Specificity.Adv Sci (Weinh). 2022 Feb;9(4):e2103645. doi: 10.1002/advs.202103645. Epub 2021 Dec 16. Adv Sci (Weinh). 2022. PMID: 34914854 Free PMC article. Review.
Cited by
-
Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933.Int J Mol Sci. 2022 Sep 1;23(17):9931. doi: 10.3390/ijms23179931. Int J Mol Sci. 2022. PMID: 36077327 Free PMC article.
-
Interactome-Seq: A Protocol for Domainome Library Construction, Validation and Selection by Phage Display and Next Generation Sequencing.J Vis Exp. 2018 Oct 3;(140):56981. doi: 10.3791/56981. J Vis Exp. 2018. PMID: 30346377 Free PMC article.
-
Selections for constituting new RNA-protein interactions in catalytic RNP.Nucleic Acids Res. 2003 Jan 15;31(2):661-9. doi: 10.1093/nar/gkg140. Nucleic Acids Res. 2003. PMID: 12527775 Free PMC article.
-
Fluorescent T7 display phages obtained by translational frameshift.Nucleic Acids Res. 2006;34(20):e137. doi: 10.1093/nar/gkl600. Epub 2006 Oct 13. Nucleic Acids Res. 2006. PMID: 17040895 Free PMC article.
-
Display technologies: application for the discovery of drug and gene delivery agents.Adv Drug Deliv Rev. 2006 Dec 30;58(15):1622-54. doi: 10.1016/j.addr.2006.09.018. Epub 2006 Oct 6. Adv Drug Deliv Rev. 2006. PMID: 17123658 Free PMC article. Review.
References
-
- Mattaj I W. Cell. 1993;73:837–840. - PubMed
-
- Burd C G, Dreyfuss G. Science. 1994;265:615–621. - PubMed
-
- Siomi H, Dreyfuss G. Curr Opin Genet Dev. 1997;7:345–353. - PubMed
-
- Ashley C T, Wilkinson K D, Reines D, Warren S T. Science. 1993;262:563–566. - PubMed
-
- Musunuru K, Darnell R B. Annu Rev Neurosci. 2001;24:239–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

