In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants
- PMID: 11679356
- PMCID: PMC93301
- DOI: 10.1128/AEM.67.11.5273-5284.2001
In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants
Abstract
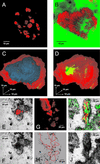
Uncultivated Nitrospira-like bacteria in different biofilm and activated-sludge samples were investigated by cultivation-independent molecular approaches. Initially, the phylogenetic affiliation of Nitrospira-like bacteria in a nitrifying biofilm was determined by 16S rRNA gene sequence analysis. Subsequently, a phylogenetic consensus tree of the Nitrospira phylum including all publicly available sequences was constructed. This analysis revealed that the genus Nitrospira consists of at least four distinct sublineages. Based on these data, two 16S rRNA-directed oligonucleotide probes specific for the phylum and genus Nitrospira, respectively, were developed and evaluated for suitability for fluorescence in situ hybridization (FISH). The probes were used to investigate the in situ architecture of cell aggregates of Nitrospira-like nitrite oxidizers in wastewater treatment plants by FISH, confocal laser scanning microscopy, and computer-aided three-dimensional visualization. Cavities and a network of cell-free channels inside the Nitrospira microcolonies were detected that were water permeable, as demonstrated by fluorescein staining. The uptake of different carbon sources by Nitrospira-like bacteria within their natural habitat under different incubation conditions was studied by combined FISH and microautoradiography. Under aerobic conditions, the Nitrospira-like bacteria in bioreactor samples took up inorganic carbon (as HCO(3)(-) or as CO(2)) and pyruvate but not acetate, butyrate, and propionate, suggesting that these bacteria can grow mixotrophically in the presence of pyruvate. In contrast, no uptake by the Nitrospira-like bacteria of any of the carbon sources tested was observed under anoxic or anaerobic conditions.
Figures


References
-
- Amann R I. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkeman A D C, van Elsas J D, de Bruigin F J, editors. Molecular microbial ecology manual. 3.3.6. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15.
-
- Bever J, Stein A, Teichmann H, editors. Weitergehende Abwasserreinigung. Munich, Germany: R. Oldenbourg Verlag; 1995.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

