Tissue distribution of products of the mouse decay-accelerating factor (DAF) genes. Exploitation of a Daf1 knock-out mouse and site-specific monoclonal antibodies
- PMID: 11683962
- PMCID: PMC1783297
- DOI: 10.1046/j.1365-2567.2001.01287.x
Tissue distribution of products of the mouse decay-accelerating factor (DAF) genes. Exploitation of a Daf1 knock-out mouse and site-specific monoclonal antibodies
Abstract
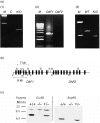
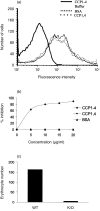
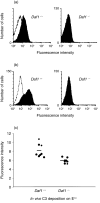
Decay-accelerating factor (DAF) is a membrane regulator of C3 activation that protects self cells from autologous complement attack. In humans, DAF is uniformly expressed as a glycosylphosphatidylinositol (GPI)-anchored molecule. In mice, both GPI-anchored and transmembrane-anchored DAF proteins are produced, each of which can be derived from two different genes (Daf1 and Daf2). In this report, we describe a Daf1 gene knock-out mouse arising as the first product of a strategy for targeting one or both Daf genes. As part of the work, we characterize recently described monoclonal antibodies against murine DAF protein using deletion mutants synthesized in yeast, and then employ the monoclonal antibodies in conjunction with wild-type and the Daf1 knock-out mice to determine the tissue distribution of the mouse Daf1 and Daf2 gene products. To enhance the immunohistochemical detection of murine DAF protein, we utilized the sensitive tyramide fluorescence method. In wild-type mice, we found strong DAF labelling of glomeruli, airway and gut epithelium, the spleen, vascular endothelium throughout all tissues, and seminiferous tubules of the testis. In Daf1 knock-out mice, DAF labelling was ablated in most tissues, but strong labelling of the testis and splenic dendritic cells remained. In both sites, reverse transcription-polymerase chain reaction analyses identified both GPI and transmembrane forms of Daf2 gene-derived protein. The results have relevance for studies of in vivo murine DAF function and of murine DAF structure.
Figures



References
-
- Nicholson-Weller A, Burge J, Fearon DT, Weller PF, Austen KF. Isolation of a human erythrocyte membrane glycoprotein with decay-accelerating activity for C3 convertases of the complement system. J Immunol. 1982;129:184–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

