Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export
- PMID: 11684705
- PMCID: PMC2150853
- DOI: 10.1083/jcb.200108007
Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export
Abstract
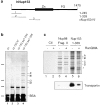
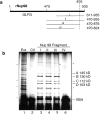
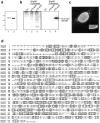
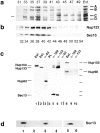
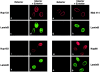
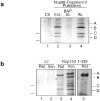
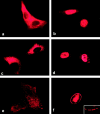
RNA undergoing nuclear export first encounters the basket of the nuclear pore. Two basket proteins, Nup98 and Nup153, are essential for mRNA export, but their molecular partners within the pore are largely unknown. Because the mechanism of RNA export will be in question as long as significant vertebrate pore proteins remain undiscovered, we set out to find their partners. Fragments of Nup98 and Nup153 were used for pulldown experiments from Xenopus egg extracts, which contain abundant disassembled nuclear pores. Strikingly, Nup98 and Nup153 each bound the same four large proteins. Purification and sequence analysis revealed that two are the known vertebrate nucleoporins, Nup96 and Nup107, whereas two mapped to ORFs of unknown function. The genes encoding the novel proteins were cloned, and antibodies were produced. Immunofluorescence reveals them to be new nucleoporins, designated Nup160 and Nup133, which are accessible on the basket side of the pore. Nucleoporins Nup160, Nup133, Nup107, and Nup96 exist as a complex in Xenopus egg extracts and in assembled pores, now termed the Nup160 complex. Sec13 is prominent in Nup98 and Nup153 pulldowns, and we find it to be a member of the Nup160 complex. We have mapped the sites that are required for binding the Nup160 subcomplex, and have found that in Nup98, the binding site is used to tether Nup98 to the nucleus; in Nup153, the binding site targets Nup153 to the nuclear pore. With transfection and in vivo transport assays, we find that specific Nup160 and Nup133 fragments block poly[A]+ RNA export, but not protein import or export. These results demonstrate that two novel vertebrate nucleoporins, Nup160 and Nup133, not only interact with Nup98 and Nup153, but themselves play a role in mRNA export.
Figures

Similar articles
-
Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex.Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):981-5. doi: 10.1073/pnas.252749899. Epub 2003 Jan 27. Proc Natl Acad Sci U S A. 2003. PMID: 12552102 Free PMC article.
-
Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr.J Cell Biol. 1998 Apr 6;141(1):31-49. doi: 10.1083/jcb.141.1.31. J Cell Biol. 1998. PMID: 9531546 Free PMC article.
-
The nucleoporin nup153 plays a critical role in multiple types of nuclear export.Mol Biol Cell. 1999 Mar;10(3):649-64. doi: 10.1091/mbc.10.3.649. Mol Biol Cell. 1999. PMID: 10069809 Free PMC article.
-
Nuclear RNA export and its importance in abiotic stress responses of plants.Curr Top Microbiol Immunol. 2008;326:235-55. doi: 10.1007/978-3-540-76776-3_13. Curr Top Microbiol Immunol. 2008. PMID: 18630756 Review.
-
Into the basket and beyond: the journey of mRNA through the nuclear pore complex.Biochem J. 2020 Jan 17;477(1):23-44. doi: 10.1042/BCJ20190132. Biochem J. 2020. PMID: 31913454 Review.
Cited by
-
ELYS regulates the localization of LBR by modulating its phosphorylation state.J Cell Sci. 2016 Nov 15;129(22):4200-4212. doi: 10.1242/jcs.190678. Epub 2016 Oct 6. J Cell Sci. 2016. PMID: 27802161 Free PMC article.
-
The nuclear pore complex and nuclear transport.Cold Spring Harb Perspect Biol. 2010 Oct;2(10):a000562. doi: 10.1101/cshperspect.a000562. Epub 2010 Jul 14. Cold Spring Harb Perspect Biol. 2010. PMID: 20630994 Free PMC article. Review.
-
The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr.Mol Biol Cell. 2005 Apr;16(4):1823-38. doi: 10.1091/mbc.e04-07-0583. Epub 2005 Jan 19. Mol Biol Cell. 2005. PMID: 15659641 Free PMC article.
-
Regulation of mRNA trafficking by nuclear pore complexes.Genes (Basel). 2014 Sep 2;5(3):767-91. doi: 10.3390/genes5030767. Genes (Basel). 2014. PMID: 25184662 Free PMC article. Review.
-
Probing nuclear pore complex architecture with proximity-dependent biotinylation.Proc Natl Acad Sci U S A. 2014 Jun 17;111(24):E2453-61. doi: 10.1073/pnas.1406459111. Epub 2014 Jun 3. Proc Natl Acad Sci U S A. 2014. PMID: 24927568 Free PMC article.
References
-
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. - PubMed
-
- Bachi, A., I.C. Braun, J.P. Rodrigues, N. Pante, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Gorlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. Rna. 6:136–158. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

