Ultrafast folding of WW domains without structured aromatic clusters in the denatured state
- PMID: 11687613
- PMCID: PMC60814
- DOI: 10.1073/pnas.221467198
Ultrafast folding of WW domains without structured aromatic clusters in the denatured state
Abstract
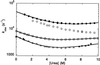
Ultrafast-folding proteins are important for combining experiment and simulation to give complete descriptions of folding pathways. The WW domain family comprises small proteins with a three-stranded antiparallel beta-sheet topology. Previous studies on the 57-residue YAP 65 WW domain indicate the presence of residual structure in the chemically denatured state. Here we analyze three minimal core WW domains of 38-44 residues. There was little spectroscopic or thermodynamic evidence for residual structure in either their chemically or thermally denatured states. Folding and unfolding kinetics, studied by using rapid temperature-jump and continuous-flow techniques, show that each domain folds and unfolds very rapidly in a two-state transition through a highly compact transition state. Folding half-times were as short as 17 micros at 25 degrees C, within an order of magnitude of the predicted maximal rate of loop formation. The small size and topological simplicity of these domains, in conjunction with their very rapid two-state folding, may allow us to reduce the difference in time scale between experiment and theoretical simulation.
Figures
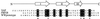
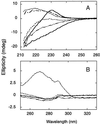
 ). The near-UV CD spectrum of free aromatic amino acids at equimolar concentrations to the aromatic content of wild-type protein, in the presence (- - - - -) and absence of 5.6 M GdmCl (– · · · –), is similar to that of the chemically denatured domain. Similar results were obtained for the prototype WW domain.
). The near-UV CD spectrum of free aromatic amino acids at equimolar concentrations to the aromatic content of wild-type protein, in the presence (- - - - -) and absence of 5.6 M GdmCl (– · · · –), is similar to that of the chemically denatured domain. Similar results were obtained for the prototype WW domain.
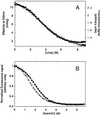
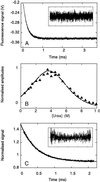
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

