COMPASS: a complex of proteins associated with a trithorax-related SET domain protein
- PMID: 11687631
- PMCID: PMC60797
- DOI: 10.1073/pnas.231473398
COMPASS: a complex of proteins associated with a trithorax-related SET domain protein
Erratum in
- Proc Natl Acad Sci U S A 2001 Dec 18;98(26):15393-4
Abstract
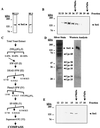
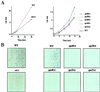
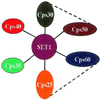
The trithorax genes encode an evolutionarily conserved family of proteins that function to maintain specific patterns of gene expression throughout cellular development. Members of this protein family contain a highly conserved 130- to 140-amino acid motif termed the SET domain. We report the purification and molecular identification of the subunits of a protein complex in the yeast Saccharomyces cerevisiae that includes the trithorax-related protein Set1. This protein complex, which we have named COMPASS (Complex Proteins Associated with Set1), consists of seven polypeptides ranging from 130 to 25 kDa. The same seven proteins were identified in COMPASS purified either by conventional biochemical chromatography or tandem-affinity tagging of the individual subunits of the complex. Null mutants missing any one of the six nonessential subunits of COMPASS grow more slowly than wild-type cells under normal conditions and demonstrate growth sensitivity to hydroxyurea. Furthermore, gene expression profiles of strains missing either of two nonessential subunits of COMPASS are altered in similar ways, suggesting these proteins have similar roles in gene expression in vivo. Molecular characterization of trithorax complexes will facilitate defining the role of this class of proteins in the regulation of gene expression and how their misregulation results in the development of human cancer.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

