Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites
- PMID: 11687644
- PMCID: PMC60849
- DOI: 10.1073/pnas.231494498
Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites
Abstract
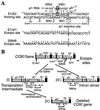
Group II introns, the presumed ancestors of nuclear pre-mRNA introns, are site-specific retroelements. In addition to "homing" to unoccupied sites in intronless alleles, group II introns transpose at low frequency to ectopic sites that resemble the normal homing site. Two general mechanisms have been proposed for group II intron transposition, one involving reverse splicing of the intron RNA directly into an ectopic DNA site, and the other involving reverse splicing into a site in RNA followed by reverse transcription and integration of the resulting cDNA by homologous recombination. Here, by using an "inverted-site" strategy, we show that the yeast mtDNA group II intron aI1 retrotransposes by reverse splicing directly into an ectopic DNA site. This same mechanism could account for other previously described ectopic transposition events in fungi and bacteria and may have contributed to the dispersal of group II introns into different genes.
Figures

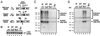


References
-
- Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. In: The RNA World. 2nd Ed. Gesteland R, Cech T R, Atkins J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 451–485.
-
- Michel F, Ferat J L. Annu Rev Biochem. 1995;64:435–461. - PubMed
-
- Belfort M, Derbyshire V, Parker M M, Cousineau B, Lambowitz A M. In: Mobile DNA. 2nd Ed. Craig N L, Craigie R, Gellert M, Lambowitz A M, editors. Washington, DC: Am. Soc. Microbiol.; 2001. , in press.
-
- Eskes R, Yang J, Lambowitz A, Perlman P. Cell. 1997;88:865–874. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

