Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element
- PMID: 11687653
- PMCID: PMC60809
- DOI: 10.1073/pnas.241286698
Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element
Abstract
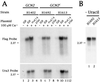
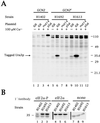
Internal initiation of translation can be mediated by specific internal ribosome entry site (IRES) elements that are located in certain mammalian and viral mRNA molecules. Thus far, these mammalian cellular and viral IRES elements have not been shown to function in the yeast Saccharomyces cerevisiae. We report here that a recently discovered IRES located in the genome of cricket paralysis virus can direct the efficient translation of a second URA3 cistron in dicistronic mRNAs in S. cerevisiae, thereby conferring uracil-independent growth. Curiously, the IRES functions poorly in wild-type yeast but functions efficiently either in the presence of constitutive expression of the eIF2 kinase GCN2 or in cells that have two initiator tRNA(met) genes disrupted. Both of these conditions have been shown to lower the amounts of ternary eIF2-GTP/initiator tRNA(met) complexes. Furthermore, tRNA(met)-independent initiation was also observed in translation-competent extracts prepared from S. cerevisiae in the presence of edeine, a compound that has been shown to interfere with start codon recognition by ribosomal subunits carrying ternary complexes. Therefore, the cricket paralysis virus IRES is likely to recruit ribosomes by internal initiation in S. cerevisiae in the absence of eIF2 and initiator tRNA(met), by the same mechanism of factor-independent ribosome recruitment used in mammalian cells. These findings will allow the use of yeast genetics to determine the mechanism of internal ribosome entry.
Figures




Comment in
-
Unleashing yeast genetics on a factor-independent mechanism of internal translation initiation.Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):12866-8. doi: 10.1073/pnas.241517998. Proc Natl Acad Sci U S A. 2001. PMID: 11698676 Free PMC article. No abstract available.
References
-
- Donahue T F. In: Translational Control of Gene Expression. Sonenberg N, Hershey J W B, Mathews M B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 487–502.
-
- Evstafieva A G, Beletsky A V, Borovjagin A V, Bogdanov A A. FEBS Lett. 1993;335:273–276. - PubMed
-
- Das S, Ott M, Yamane A, Venkatesan A, Gupta S, Dasgupta A. Front Biosci. 1998;3:D1241–D1252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

