The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein
- PMID: 11689427
- PMCID: PMC125290
- DOI: 10.1093/emboj/20.21.5863
The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein
Abstract
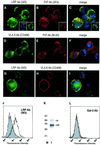
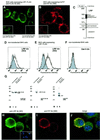
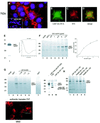
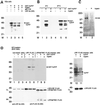
Recently, we identified the 37-kDa laminin receptor precursor (LRP) as an interactor for the prion protein (PrP). Here, we show the presence of the 37-kDa LRP and its mature 67-kDa form termed high-affinity laminin receptor (LR) in plasma membrane fractions of N2a cells, whereas only the 37-kDa LRP was detected in baby hamster kidney (BHK) cells. PrP co-localizes with LRP/LR on the surface of N2a cells and Semliki Forest virus (SFV) RNA transfected BHK cells. Cell-binding assays reveal the LRP/LR-dependent binding of cellular PrP by neuronal and non-neuronal cells. Hyperexpression of LRP on the surface of BHK cells results in the binding of exogenous PrP. Cell binding is similar in PrP(+/+) and PrP(0/0) primary neurons, demonstrating that PrP does not act as a co-receptor of LRP/LR. LRP/LR-dependent internalization of PrP is blocked at 4 degrees C. Secretion of an LRP mutant lacking the transmembrane domain (aa 86-101) from BHK cells abolishes PrP binding and internalization. Our results show that LRP/LR acts as the receptor for cellular PrP on the surface of mammalian cells.
Figures


References
-
- Ardini E., Pesole,G., Tagliabue,E., Magnifico,A., Castronovo,V., Sobel,M.E., Colnaghi,M.I. and Menard,S. (1998) The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol. Biol. Evol., 15, 1017–1025. - PubMed
-
- Brown D.R. et al. (1997) The cellular prion protein binds copper in vivo. Nature, 390, 684–687. - PubMed
-
- Bueler H., Aguzzi,A., Sailer,A., Greiner,R.A., Autenried,P., Aguet,M. and Weissmann,C. (1993) Mice devoid of PrP are resistant to scrapie. Cell, 73, 1339–1347. - PubMed
-
- Buto S. et al. (1998) Formation of the 67-kDa laminin receptor by acylation of the precursor. J. Cell. Biochem., 69, 244–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

