The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation
- PMID: 11689437
- PMCID: PMC125692
- DOI: 10.1093/emboj/20.21.5971
The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation
Abstract
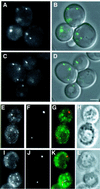
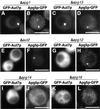
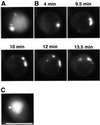
Macroautophagy is a bulk degradation process induced by starvation in eukaryotic cells. In yeast, 15 Apg proteins coordinate the formation of autophagosomes. Several key reactions performed by these proteins have been described, but a comprehensive understanding of the overall network is still lacking. Based on Apg protein localization, we have identified a novel structure that functions in autophagosome formation. This pre-autophagosomal structure, containing at least five Apg proteins, i.e. Apg1p, Apg2p, Apg5p, Aut7p/Apg8p and Apg16p, is localized in the vicinity of the vacuole. Analysis of apg mutants revealed that the formation of both a phosphatidylethanolamine-conjugated Aut7p and an Apg12p- Apg5p conjugate is essential for the localization of Aut7p to the pre-autophagosomal structure. Vps30p/Apg6p and Apg14p, components of an autophagy- specific phosphatidylinositol 3-kinase complex, Apg9p and Apg16p are all required for the localization of Apg5p and Aut7p to the structure. The Apg1p protein kinase complex functions in the late stage of autophagosome formation. Here, we present the classification of Apg proteins into three groups that reflect each step of autophagosome formation.
Figures







References
-
- Cadwell R.C. and Joyce,G.F. (1995) Mutagenic PCR. In Dieffenbach,C.W. and Dveksler,G.S. (eds), PCR Primer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 583–589.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

