Autonomous role of 3'-terminal CCCA in directing transcription of RNAs by Qbeta replicase
- PMID: 11689618
- PMCID: PMC114723
- DOI: 10.1128/JVI.75.23.11373-11383.2001
Autonomous role of 3'-terminal CCCA in directing transcription of RNAs by Qbeta replicase
Abstract
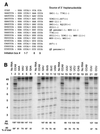
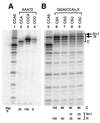
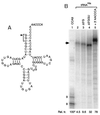
We have studied transcription in vitro by Qbeta replicase to deduce the minimal features needed for efficient end-to-end copying of an RNA template. Our studies have used templates ca. 30 nucleotides long that are expected to be free of secondary structure, permitting unambiguous analysis of the role of template sequence in directing transcription. A 3'-terminal CCCA (3'-CCCA) directs transcriptional initiation to opposite the underlined C; the amount of transcription is comparable between RNAs possessing upstream (CCA)(n) tracts, A-rich sequences, or a highly folded domain and is also comparable in single-round transcription assays to transcription of two amplifiable RNAs. Predominant initiation occurs within the 3'-CCCA initiation box when a wide variety of sequences is present immediately upstream, but CCA or a closely similar sequence in that position results in significant internal initiation. Removal of the 3'-A from the 3'-CCCA results in 5- to 10-fold-lower transcription, emphasizing the importance of the nontemplated addition of 3'-A by Qbeta replicase during termination. In considering whether 3'-CCCA could provide sufficient specificity for viral transcription, and consequently amplification, in vivo, we note that tRNA(His) is the only stable Escherichia coli RNA with 3'-CCCA. In vitro-generated transcripts corresponding to tRNA(His) served as poor templates for Qbeta replicase; this was shown to be due to the inaccessibility of the partially base-paired CCCA. These studies demonstrate that 3'-CCCA plays a major role in the control of transcription by Qbeta replicase and that the abundant RNAs present in the host cell should not be efficient templates.
Figures
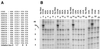



References
-
- Arora R, Priano C C, Jacobson A B, Mills D R. cis-Acting elements within an RNA coliphage genome: fold as you please, but fold you must! J Mol Biol. 1996;258:433–446. - PubMed
-
- Avota E, Berzins V, Grens E, Vishnevsky Y, Luce R, Biebricher C K. The natural 6S RNA found in Qβ-infected cells is derived from host and phage RNA. J Mol Biol. 1998;276:7–17. - PubMed
-
- Beekwilder M J, Nieuwenhuizen R, van Duin J. Secondary structure model for the last two domains of single-stranded RNA phage Qβ. J Mol Biol. 1995;247:903–917. - PubMed
-
- Biebricher C K, Luce R. Sequence analysis of RNA species synthesized by Qβ replicase without template. Biochemistry. 1993;32:4848–4854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

