Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication
- PMID: 11689621
- PMCID: PMC114726
- DOI: 10.1128/JVI.75.23.11401-11407.2001
Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication
Abstract
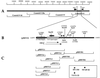
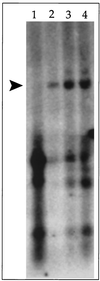
We have identified a lytic origin of DNA replication (oriLyt) for rhesus macaque rhadinovirus (RRV), the rhesus macaque homolog of human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma-associated herpesvirus. RRV oriLyt maps to the region of the genome between open reading frame 69 (ORF69) and ORF71 (vFLIP) and is composed of an upstream A+T-rich region followed by a short (300-bp) downstream G+C-rich DNA sequence. A set of overlapping cosmids corresponding to the entire genome of RRV was capable of complementing oriLyt-dependent DNA replication only when additional ORF50 was supplied as an expression plasmid in the transfection mixture, suggesting that the level of ORF50 protein originating from input cosmid DNA was insufficient. The requirement of RRV ORF50 in the cotransfection replication assay may also suggest a direct role for this protein in DNA replication. RRV oriLyt shares a high degree of nucleotide sequence and G+C base distribution with the corresponding loci in HHV-8.
Figures



Similar articles
-
A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization.J Virol. 2006 Feb;80(3):1549-62. doi: 10.1128/JVI.80.3.1549-1562.2006. J Virol. 2006. PMID: 16415030 Free PMC article.
-
A G protein-coupled receptor encoded by rhesus rhadinovirus is similar to ORF74 of Kaposi's sarcoma-associated herpesvirus.J Virol. 2003 Feb;77(3):1738-46. doi: 10.1128/jvi.77.3.1738-1746.2003. J Virol. 2003. PMID: 12525607 Free PMC article.
-
Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes.J Virol. 2002 Oct;76(19):9819-31. doi: 10.1128/jvi.76.19.9819-9831.2002. J Virol. 2002. PMID: 12208960 Free PMC article.
-
Rhesus monkey rhadinovirus: a model for the study of KSHV.Curr Top Microbiol Immunol. 2007;312:43-69. doi: 10.1007/978-3-540-34344-8_2. Curr Top Microbiol Immunol. 2007. PMID: 17089793 Review.
-
Rhesus macaque rhadinovirus-associated disease.Curr Opin Virol. 2013 Jun;3(3):245-50. doi: 10.1016/j.coviro.2013.05.016. Epub 2013 Jun 6. Curr Opin Virol. 2013. PMID: 23747119 Free PMC article. Review.
Cited by
-
Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68.J Virol. 2004 Sep;78(17):9123-31. doi: 10.1128/JVI.78.17.9123-9131.2004. J Virol. 2004. PMID: 15308708 Free PMC article.
-
Multiple Lytic Origins of Replication Are Required for Optimal Gammaherpesvirus Fitness In Vitro and In Vivo.PLoS Pathog. 2016 Mar 23;12(3):e1005510. doi: 10.1371/journal.ppat.1005510. eCollection 2016 Mar. PLoS Pathog. 2016. PMID: 27007137 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication.J Virol. 2002 Aug;76(15):7890-6. doi: 10.1128/jvi.76.15.7890-7896.2002. J Virol. 2002. PMID: 12097603 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin.J Virol. 2003 May;77(10):5578-88. doi: 10.1128/jvi.77.10.5578-5588.2003. J Virol. 2003. PMID: 12719550 Free PMC article.
-
Evaluation of the lytic origins of replication of Kaposi's sarcoma-associated virus/human herpesvirus 8 in the context of the viral genome.J Virol. 2006 Oct;80(19):9905-9. doi: 10.1128/JVI.01004-06. J Virol. 2006. PMID: 16973596 Free PMC article.
References
-
- Birley H D, Schultz T F. Kaposi's sarcoma and the new herpesvirus. J Med Microbiol. 1997;46:433–435. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

