Identification, phylogeny, and evolution of retroviral elements based on their envelope genes
- PMID: 11689652
- PMCID: PMC114757
- DOI: 10.1128/JVI.75.23.11709-11719.2001
Identification, phylogeny, and evolution of retroviral elements based on their envelope genes
Abstract
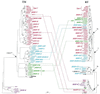
Phylogenetic analyses of retroviral elements, including endogenous retroviruses, have relied essentially on the retroviral pol gene expressing the highly conserved reverse transcriptase. This enzyme is essential for the life cycle of all retroid elements, but other genes are also endowed with conserved essential functions. Among them, the transmembrane (TM) subunit of the envelope gene is involved in virus entry through membrane fusion. It has also been reported to contain a domain, named the immunosuppressive domain, that has immunosuppressive properties most probably essential for virus spread within the host. This domain is conserved among a large series of retroviral elements, and we have therefore attempted to generate phylogenetic links between retroviral elements identified from databases following tentative alignments of the immunosuppressive domain and adjacent sequences. This allowed us to unravel a conserved organization among TM domains, also found in the Ebola and Marburg filoviruses, and to identify a large number of human endogenous retroviruses (HERVs) from sequence databases. The latter elements are part of previously identified families of HERVs, and some of them define new families. A general phylogenetic analysis based on the TM proteins of retroelements, and including those with no clearly identified immunosuppressive domain, could then be derived and compared with pol-based phylogenetic trees, providing a comprehensive survey of retroelements and definitive evidence for recombination events in the generation of both the endogenous and the present-day infectious retroviruses.
Figures




References
-
- Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. - PubMed
-
- Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

