Interaction of zyxin, a focal adhesion protein, with the e6 protein from human papillomavirus type 6 results in its nuclear translocation
- PMID: 11689660
- PMCID: PMC114765
- DOI: 10.1128/JVI.75.23.11791-11802.2001
Interaction of zyxin, a focal adhesion protein, with the e6 protein from human papillomavirus type 6 results in its nuclear translocation
Abstract
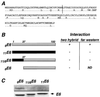
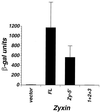
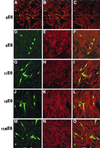
Zyxin, a focal adhesion molecule, interacts specifically with the E6 protein from human papillomavirus (HPV) type 6 in a yeast two-hybrid screen of a cDNA library prepared from human keratinocytes. Zyxin does not interact significantly with E6 proteins from HPV types 11, 16, or 18. The interaction was confirmed by in vitro and in vivo analyses and it requires the LIM domains (Lin-11, Isl-1, and Mec-3 [G. Freyd, S. K. Kim, and H. R. Horvitz, Nature 344:876-879, 1990]) found at the carboxyl terminus of zyxin. Cotransfection of E6 from HPV ((6)E6) and zyxin results in the accumulation of zyxin in the nucleus where it can function as a transcriptional activator. (6)E6 can also mobilize endogenous zyxin to the nucleus.
Figures
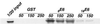





Similar articles
-
Targeting of zyxin to sites of actin membrane interaction and to the nucleus.J Biol Chem. 2001 Sep 14;276(37):34759-67. doi: 10.1074/jbc.M102820200. Epub 2001 Jun 6. J Biol Chem. 2001. PMID: 11395501
-
SH3 domain-dependent interaction of the proto-oncogene product Vav with the focal contact protein zyxin.Oncogene. 1996 Apr 4;12(7):1577-81. Oncogene. 1996. PMID: 8622875
-
The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300.EMBO J. 1999 Sep 15;18(18):5061-72. doi: 10.1093/emboj/18.18.5061. EMBO J. 1999. PMID: 10487758 Free PMC article.
-
Zyxin: zinc fingers at sites of cell adhesion.Bioessays. 1997 Nov;19(11):949-57. doi: 10.1002/bies.950191104. Bioessays. 1997. PMID: 9394617 Review.
-
Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear.Biochim Biophys Acta. 2003 Feb 17;1593(2-3):115-20. doi: 10.1016/s0167-4889(02)00349-x. Biochim Biophys Acta. 2003. PMID: 12581855 Review.
Cited by
-
The role of zyxin in regulation of malignancies.Heliyon. 2018 Jul 18;4(7):e00695. doi: 10.1016/j.heliyon.2018.e00695. eCollection 2018 Jul. Heliyon. 2018. PMID: 30094365 Free PMC article. Review.
-
Zyxin (ZYX) promotes invasion and acts as a biomarker for aggressive phenotypes of human glioblastoma multiforme.Lab Invest. 2020 Jun;100(6):812-823. doi: 10.1038/s41374-019-0368-9. Epub 2020 Jan 16. Lab Invest. 2020. PMID: 31949244
-
Zyxin regulates migration of renal epithelial cells through activation of hepatocyte nuclear factor-1β.Am J Physiol Renal Physiol. 2013 Jul 1;305(1):F100-10. doi: 10.1152/ajprenal.00582.2012. Epub 2013 May 8. Am J Physiol Renal Physiol. 2013. PMID: 23657850 Free PMC article.
-
Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast.Breast Cancer Res. 2005;7(1):R1-11. doi: 10.1186/bcr940. Epub 2004 Oct 22. Breast Cancer Res. 2005. PMID: 15642157 Free PMC article.
-
The role of zyxin in signal transduction and its relationship with diseases.Front Mol Biosci. 2024 Apr 22;11:1371549. doi: 10.3389/fmolb.2024.1371549. eCollection 2024. Front Mol Biosci. 2024. PMID: 38712343 Free PMC article. Review.
References
-
- Arber S, Hunter J J, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard J C, Chien K R, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. - PubMed
-
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

