Molecular evolution of Puumala hantavirus
- PMID: 11689661
- PMCID: PMC114766
- DOI: 10.1128/JVI.75.23.11803-11810.2001
Molecular evolution of Puumala hantavirus
Abstract
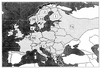
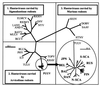
Puumala virus (PUUV) is a negative-stranded RNA virus in the genus Hantavirus, family Bunyaviridae. In this study, detailed phylogenetic analysis was performed on 42 complete S segment sequences of PUUV originated from several European countries, Russia, and Japan, the largest set available thus far for hantaviruses. The results show that PUUV sequences form seven distinct and well-supported genetic lineages; within these lineages, geographical clustering of genetic variants is observed. The overall phylogeny of PUUV is star-like, suggesting an early split of genetic lineages. The individual PUUV lineages appear to be independent, with the only exception to this being the Finnish and the Russian lineages that are closely connected to each other. Two strains of PUUV-like virus from Japan form the most ancestral lineage diverging from PUUV. Recombination points within the S segment were searched for and evidence for intralineage recombination events was seen in the Finnish, Russian, Danish, and Belgian lineages of PUUV. Molecular clock analysis showed that PUUV is a stable virus, evolving slowly at a rate of 0.7 x 10(-7) to 2.2 x 10(-6) nt substitutions per site per year.
Figures
References
-
- Aberle S W, Lehner P, Ecker M, Aberle J H, Arneitz K, Khanakah G, Radda A, Radda I, Popow-Kraupp T, Kunz C, Heinz F X. Nephropathia epidemica and Puumala virus in Austria. Eur J Clin Microbiol Infect Dis. 1999;18:467–472. - PubMed
-
- Asikainen K, Hänninen T, Henttonen H, Niemimaa J, Laakkonen J, Andersen H K, Bille N, Leirs H, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus in Fennoscandia: phylogenetic analysis of strains from two recolonization routes, Karelia and Denmark. J Gen Virol. 2000;81:2833–2841. - PubMed
-
- Bandelt H-J, Dress A W M. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. - PubMed
-
- Bowen M D, Gelbmann W, Ksiazek T G, Nichol S T, Nowotny N. Puumala virus and two genetic variants of Tula virus are present in Austrian rodents. J Med Virol. 1997;53:174–181. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

